TCRcloud is an AIRR visualization and comparison tool
Project description





TCRcloud is an Adaptive Immune Receptor Repertoire (AIRR) visualization and comparison tool
Instalation
TCRcloud is written in python and can be installed from PyPI using pip:
pip3 install TCRcloud
It is compatible with Linux and macOS operating systems and on Windows through the Windows Subsystem for Linux.
TCRcloud uses the AIRR Data Commons API and needs AIRR compliant data as input.
TCRcloud was initially developed for TCR repertoires but it is also compatible with BCR repertoires.
Usage
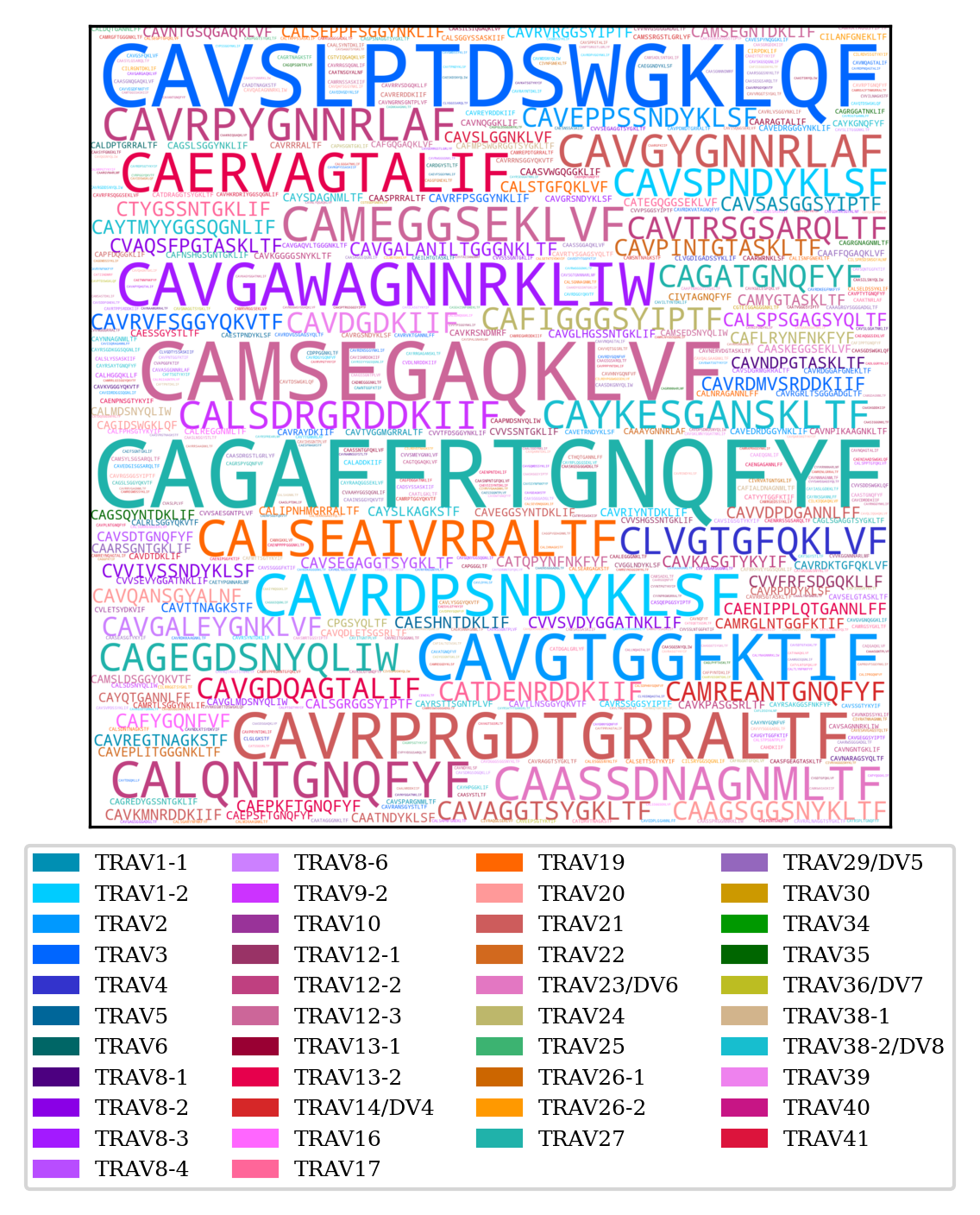
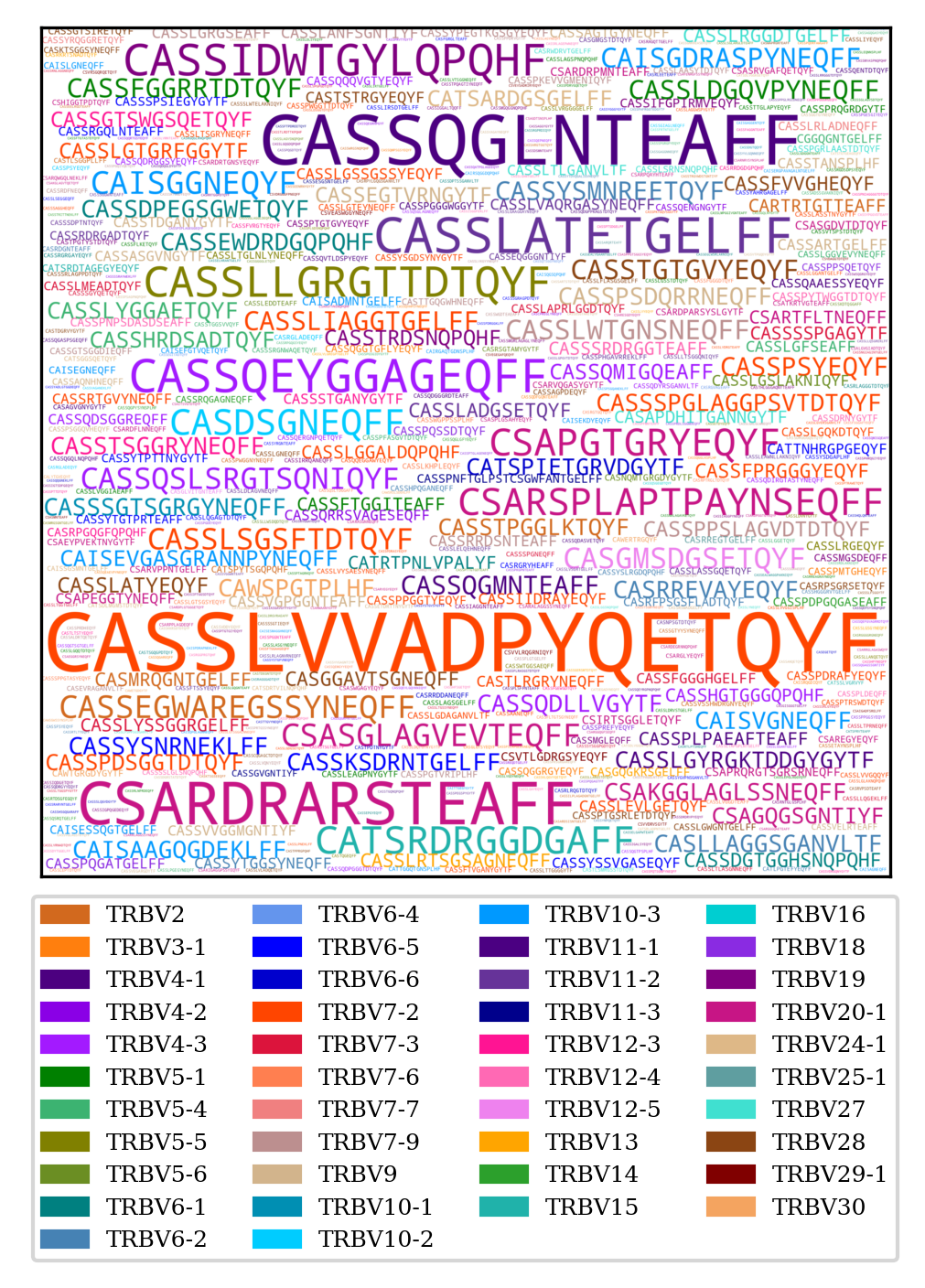
To create a word cloud
TCRcloud cloud repertoire.airr.rearrangements.tsv
By default TCRcloud colours the CDR3 based on the V gene. Only the colours for human TCR and BCR variable genes are coded into TCRcloud but you can provide a json file that atributes colours in Hex format to specific sequences:
{
"#FF0000":["CAVSLPTDSWGKLQF","CASSLVVADPYQETQYF"],
"#0000FF":["CAYRSKGSQGNLIF","CASSLGGQSGNEQFF"]
}
The sequences not in the json file will be coloured grey.
To use your custom colours for the word cloud
TCRcloud cloud repertoire.airr.rearrangements.tsv -c colours.json
To create a word cloud without a legend
TCRcloud cloud repertoire.airr.rearrangements.tsv -l False
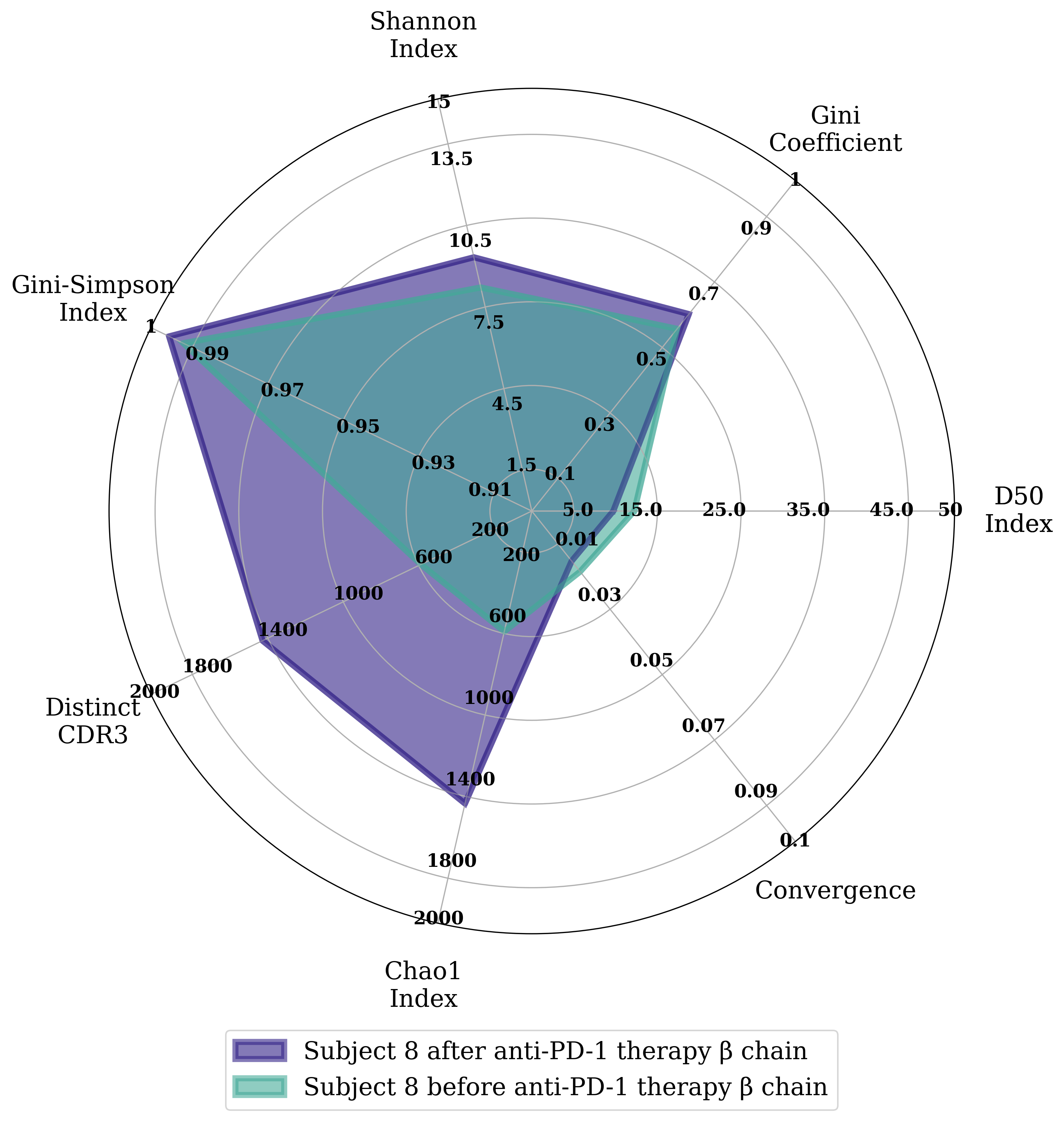
To create a radar plot comparing diversity indices
TCRcloud radar repertoire.airr.rearrangements.tsv
By default TCRcloud uses repertoire_id but you can create a legend with the text you want by providing a json file:
{
"PRJNA509910-su008_pre-TRA":"Subject 8 pre-treatment",
"PRJNA509910-su008_post-TRA":"Subject 8 post-treatment",
"PRJNA509910-su008_pre-TRB":"Subject 8 pre-treatment",
"PRJNA509910-su008_post-TRB":"Subject 8 post-treatment"
}
To create a radar plot with your desired legend
TCRcloud radar repertoire.airr.rearrangements.tsv -c legend.json
To create a radar plot without a legend
TCRcloud radar repertoire.airr.rearrangements.tsv -l False
To export the calculated indices from the radar to a text file
TCRcloud radar repertoire.airr.rearrangements.tsv -e True
The indices utilized in the radar
Distinct CDR3: Count of the unique CDR3 sequences in the sample
Convergence: Frequency of CDR3 amino acid sequences that are coded by more than one nucleotide sequence
D50 Index: Developed by Dr Jian Han, this index represents the percent of dominant and unique T or B cell clones that account for the cumulative 50% of the total CDR3 counted in the sample
Gini Coefficient: Originally developed by Dr Corrado Gini to represent the wealth inequality within a social group
Shannon Index: Originally developed by Dr Claude Shannon to quantify the entropy in strings of text. Calculated here using log2
Gini-Simpson Index: This is a transformation of the index originally developed by Dr Edward H. Simpson to measure diversity in a ecosystem
Chao1 index: Originally developed by Dr Anne Chao to estimate richness in a ecological community
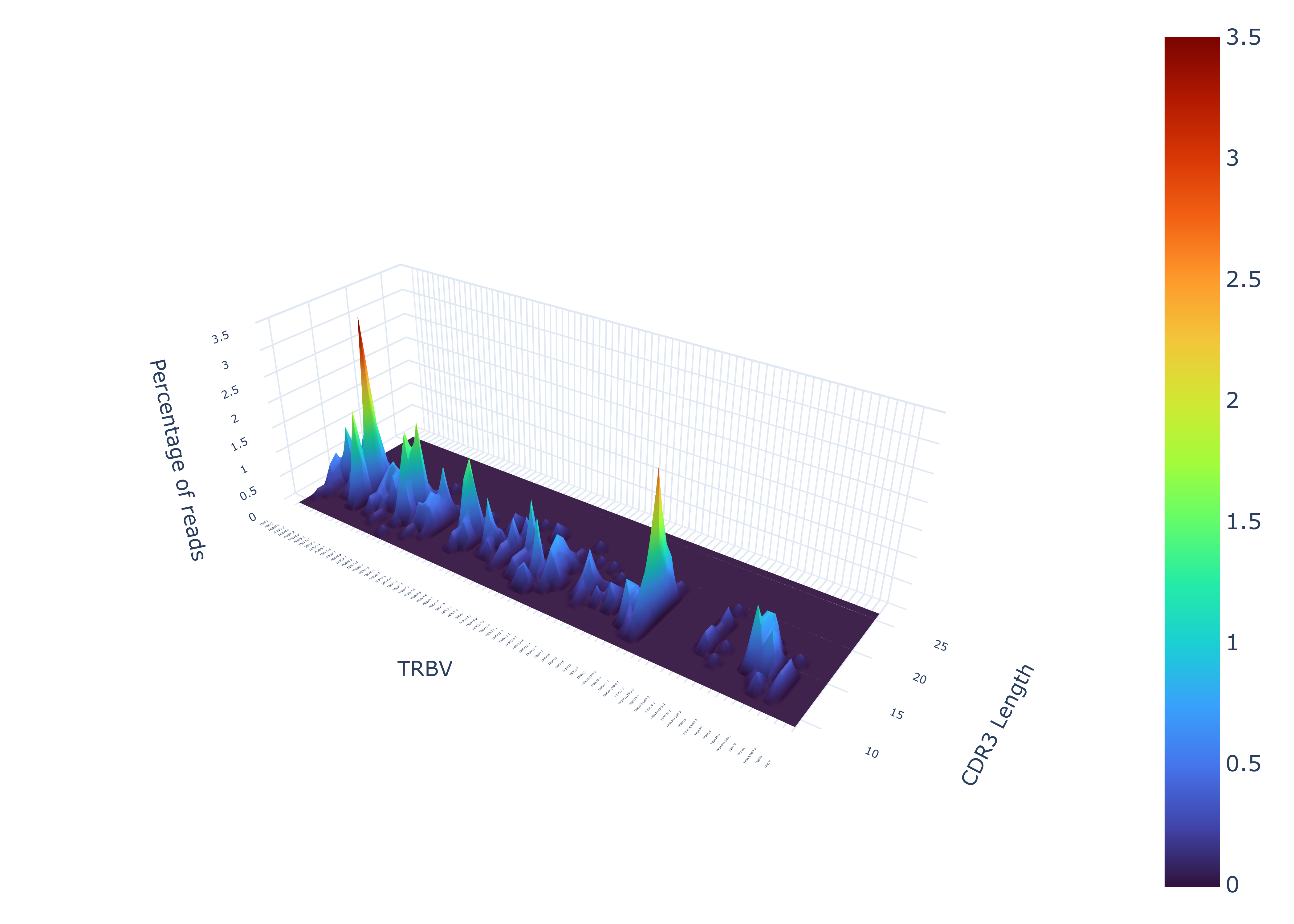
To create an amino V-genes plot
TCRcloud vgenes repertoire.airr.rearrangements.tsv
To export the processed data from the V-genes plot to a text file
TCRcloud vgenes repertoire.airr.rearrangements.tsv -e True
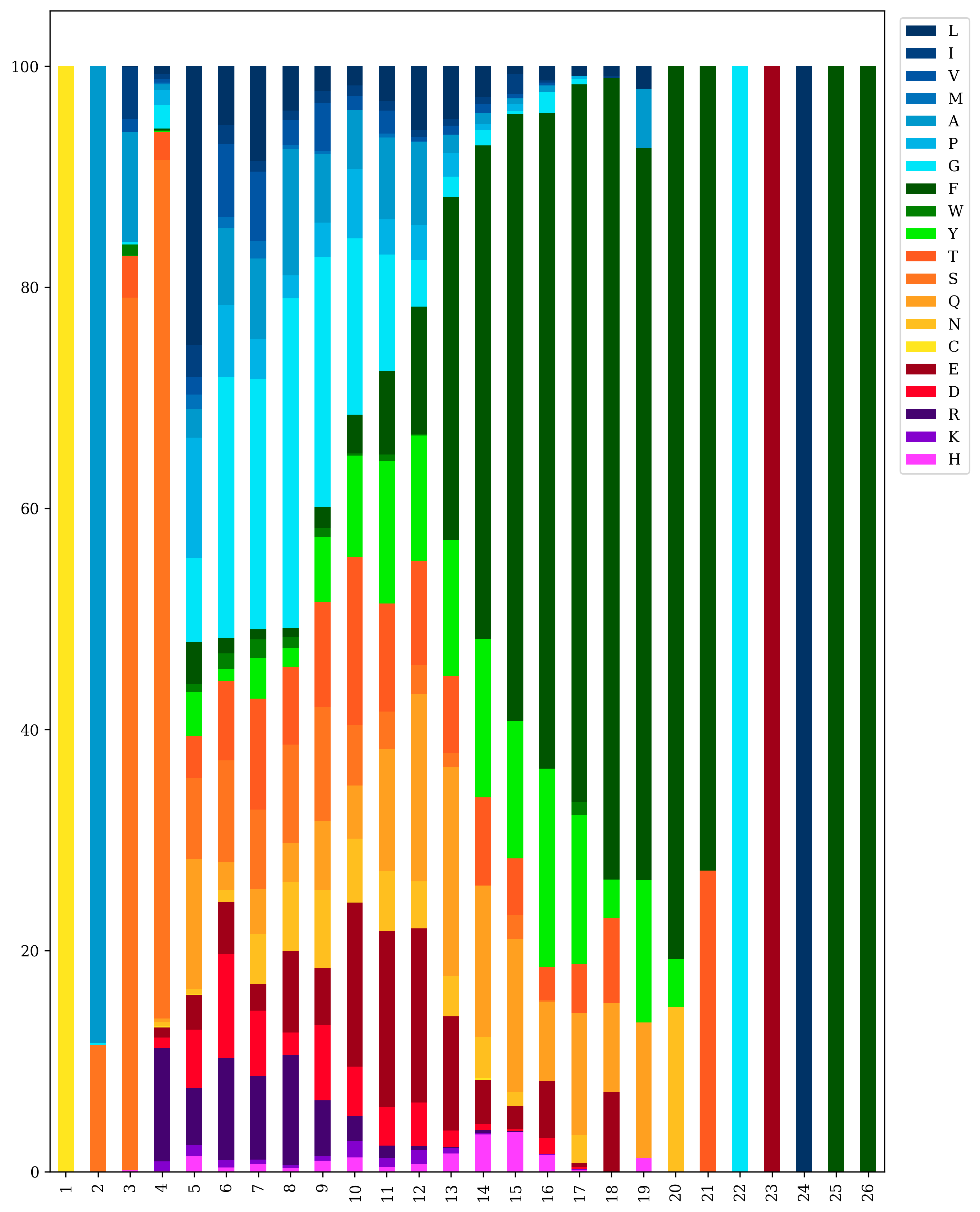
To create a 2-D amino acid plot
TCRcloud aminoacids repertoire.airr.rearrangements.tsv
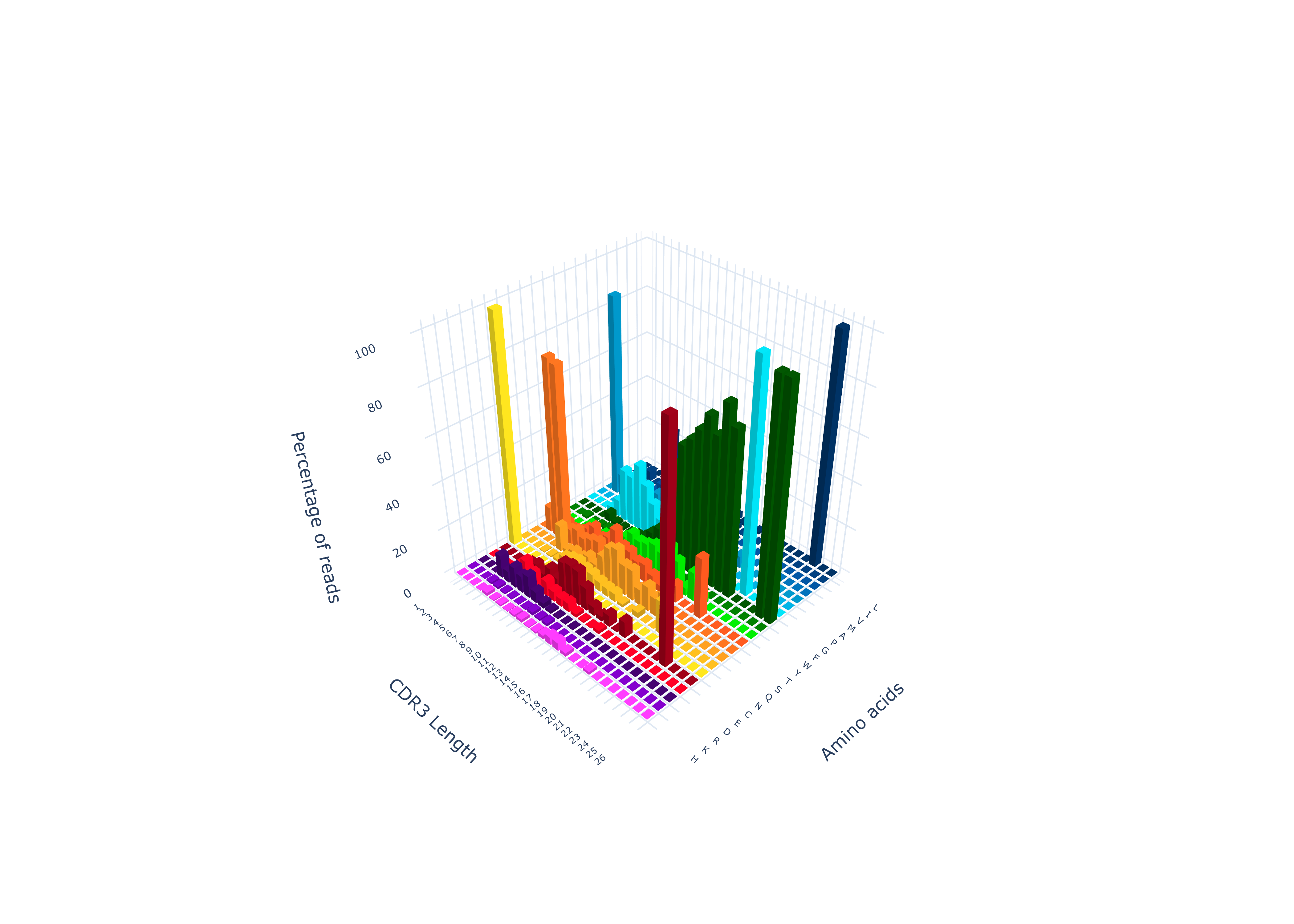
To create a 3-D amino acid plot
TCRcloud aminoacids repertoire.airr.rearrangements.tsv -t True
To export the processed data from the amino acid plot to a text file
TCRcloud aminoacids repertoire.airr.rearrangements.tsv -e True
Using TCRcloud you can download rearragements files from the AIRR compliant databases based on AIRR repertoire metadata files
To download AIRR rearrangements files
TCRcloud download repertoire.airr.json
TCRcloud provides some test data to experiment the tool. The data is from subject number 8 of Yost et al. (2019) (DOI: 10.1038/s41591-019-0522-3)
To download the test data repertoire file
TCRcloud testdata
After having the alpharepertoire.airr.json and betarepertoire.airr.json file you can use the download function included in TCRcloud to get the matching rearragements file.
Examples:
TRA CDR3 word cloud
TRB CDR3 word cloud
Diversity comparison
V-genes plot
2-D Amino acid plot
3-D Amino acid plot
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file TCRcloud-1.5.1.tar.gz.
File metadata
- Download URL: TCRcloud-1.5.1.tar.gz
- Upload date:
- Size: 22.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.12.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
4712a1624a844124b5e4bc58231c483531e139c1d66c6ee192d5c34210557cd9
|
|
| MD5 |
b0b3ce83e0d35f7d8bee1ca8637c66a0
|
|
| BLAKE2b-256 |
baf55718e336320e91de5d72514f0bab12733c20799b4199e6034a5a11732bd2
|
File details
Details for the file TCRcloud-1.5.1-py3-none-any.whl.
File metadata
- Download URL: TCRcloud-1.5.1-py3-none-any.whl
- Upload date:
- Size: 25.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.12.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b36db6945b868c94354d9df0bb3e01f16d406ea333d1886ca3ff35bbabc7e74c
|
|
| MD5 |
bbe764fa05ed347d4cb2a16c65dd0515
|
|
| BLAKE2b-256 |
11516625ff4355cc83b44220c436ba2e7d166009d8cf3a4318dfde8b4804c90d
|