A framework for Bayesian multi-object tracking
Project description
Bayesian Tracker (btrack) 🔬💻
btrack is a Python library for multi object tracking, used to reconstruct trajectories in crowded fields.
Here, we use a probabilistic network of information to perform the trajectory linking.
This method uses spatial information as well as appearance information for track linking.
The tracking algorithm assembles reliable sections of track that do not contain splitting events (tracklets). Each new tracklet initiates a probabilistic model, and utilises this to predict future states (and error in states) of each of the objects in the field of view. We assign new observations to the growing tracklets (linking) by evaluating the posterior probability of each potential linkage from a Bayesian belief matrix for all possible linkages.
The tracklets are then assembled into tracks by using multiple hypothesis testing and integer programming to identify a globally optimal solution. The likelihood of each hypothesis is calculated for some or all of the tracklets based on heuristics. The global solution identifies a sequence of high-likelihood hypotheses that accounts for all observations.
We developed btrack for cell tracking in time-lapse microscopy data.
Installation
btrack has been tested with 
x86_64 macos>=11, ubuntu>=20.04 and windows>=10.0.17763.
Note that btrack<=0.5.0 was built against earlier version of
Eigen which used C++=11, as of btrack==0.5.1
it is now built against C++=17.
Installing the latest stable version
pip install btrack
Usage examples
Visit btrack documentation to learn how to use it and see other examples.
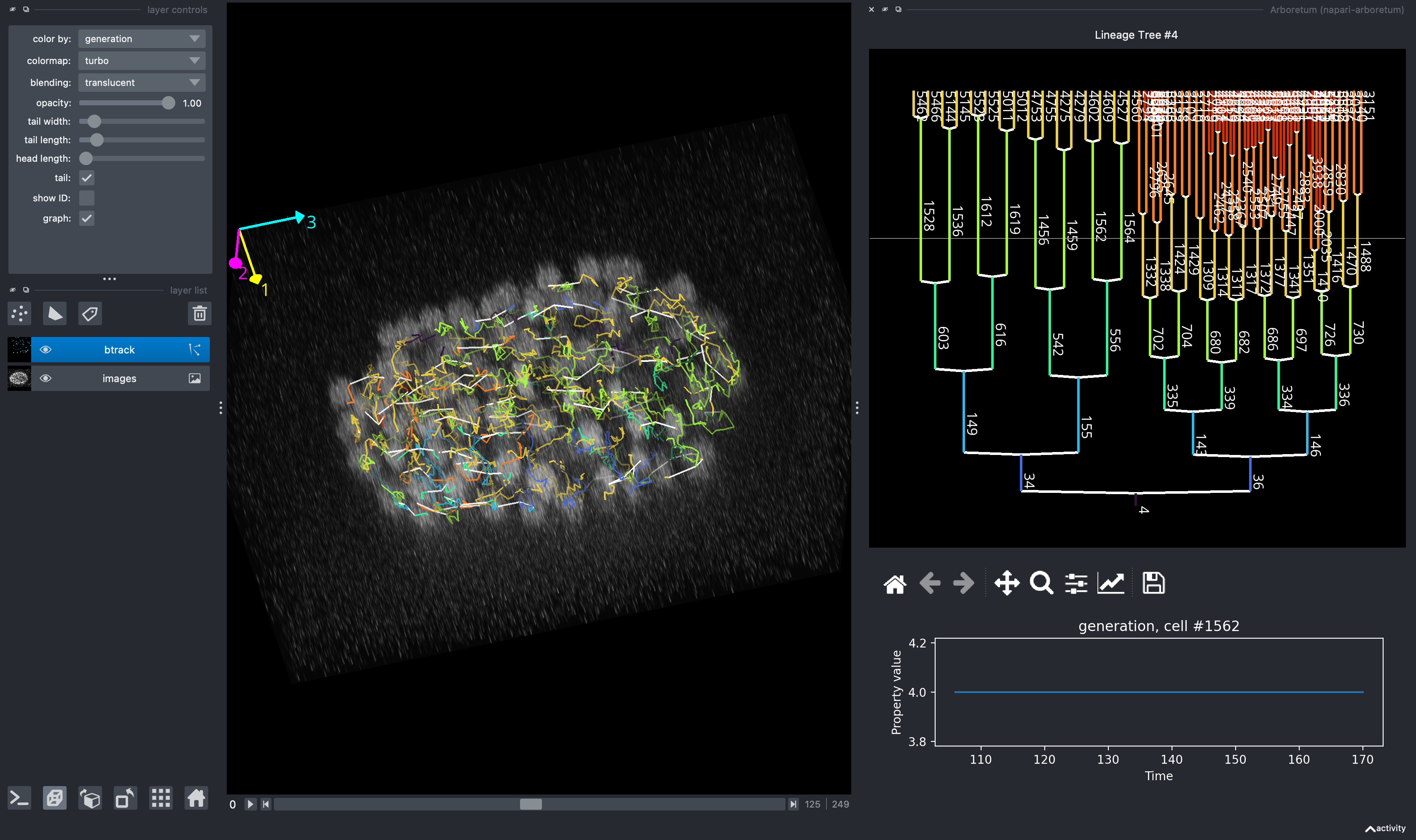
Cell tracking in time-lapse imaging data
We provide integration with Napari, including a plugin for graph visualization, arboretum.
Video of tracking, showing automatic lineage determination
Development
The tracker and hypothesis engine are mostly written in C++ with a Python wrapper. If you would like to contribute to btrack, you will need to install the latest version from GitHub. Follow the instructions on our developer guide.
Citation
More details of how this type of tracking approach can be applied to tracking cells in time-lapse microscopy data can be found in the following publications:
Automated deep lineage tree analysis using a Bayesian single cell tracking approach
Ulicna K, Vallardi G, Charras G and Lowe AR.
Front in Comp Sci (2021)
Local cellular neighbourhood controls proliferation in cell competition
Bove A, Gradeci D, Fujita Y, Banerjee S, Charras G and Lowe AR.
Mol. Biol. Cell (2017)
@ARTICLE {10.3389/fcomp.2021.734559,
AUTHOR = {Ulicna, Kristina and Vallardi, Giulia and Charras, Guillaume and Lowe, Alan R.},
TITLE = {Automated Deep Lineage Tree Analysis Using a Bayesian Single Cell Tracking Approach},
JOURNAL = {Frontiers in Computer Science},
VOLUME = {3},
PAGES = {92},
YEAR = {2021},
URL = {https://www.frontiersin.org/article/10.3389/fcomp.2021.734559},
DOI = {10.3389/fcomp.2021.734559},
ISSN = {2624-9898}
}
@ARTICLE {Bove07112017,
author = {Bove, Anna and Gradeci, Daniel and Fujita, Yasuyuki and Banerjee,
Shiladitya and Charras, Guillaume and Lowe, Alan R.},
title = {Local cellular neighborhood controls proliferation in cell competition},
volume = {28},
number = {23},
pages = {3215-3228},
year = {2017},
doi = {10.1091/mbc.E17-06-0368},
URL = {http://www.molbiolcell.org/content/28/23/3215.abstract},
eprint = {http://www.molbiolcell.org/content/28/23/3215.full.pdf+html},
journal = {Molecular Biology of the Cell}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file btrack-0.7.0.tar.gz.
File metadata
- Download URL: btrack-0.7.0.tar.gz
- Upload date:
- Size: 1.3 MB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
56f29ba3e0b68bc6c87f2e4a6bbdf2cc8c0ca75f21d0caabf3801299976da782
|
|
| MD5 |
4d0d5d4f06f3e3c99bd1d05b444b92a3
|
|
| BLAKE2b-256 |
8d59dc0578d7778861f64fbf659e8f786de7f4dbb9d85b4851c2c0fd11e3791e
|
Provenance
The following attestation bundles were made for btrack-0.7.0.tar.gz:
Publisher:
deploy.yml on quantumjot/btrack
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
btrack-0.7.0.tar.gz -
Subject digest:
56f29ba3e0b68bc6c87f2e4a6bbdf2cc8c0ca75f21d0caabf3801299976da782 - Sigstore transparency entry: 567410828
- Sigstore integration time:
-
Permalink:
quantumjot/btrack@a3bd947915efe6837936f9db6db88417f0b51b45 -
Branch / Tag:
refs/tags/v0.7.0 - Owner: https://github.com/quantumjot
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
deploy.yml@a3bd947915efe6837936f9db6db88417f0b51b45 -
Trigger Event:
push
-
Statement type:
File details
Details for the file btrack-0.7.0-py3-none-any.whl.
File metadata
- Download URL: btrack-0.7.0-py3-none-any.whl
- Upload date:
- Size: 1.3 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
1144abc5419ab382a6dc84e593c3d8930b92cf118eae450255298ebc378eaa23
|
|
| MD5 |
0c97b8f2451dd6a15802d7a9734bd693
|
|
| BLAKE2b-256 |
8197135e95fc59cd892dbdbeb890d85ac139bbfde41e618dbb7cbe7bb1c208d6
|
Provenance
The following attestation bundles were made for btrack-0.7.0-py3-none-any.whl:
Publisher:
deploy.yml on quantumjot/btrack
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
btrack-0.7.0-py3-none-any.whl -
Subject digest:
1144abc5419ab382a6dc84e593c3d8930b92cf118eae450255298ebc378eaa23 - Sigstore transparency entry: 567410833
- Sigstore integration time:
-
Permalink:
quantumjot/btrack@a3bd947915efe6837936f9db6db88417f0b51b45 -
Branch / Tag:
refs/tags/v0.7.0 - Owner: https://github.com/quantumjot
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
deploy.yml@a3bd947915efe6837936f9db6db88417f0b51b45 -
Trigger Event:
push
-
Statement type:






















