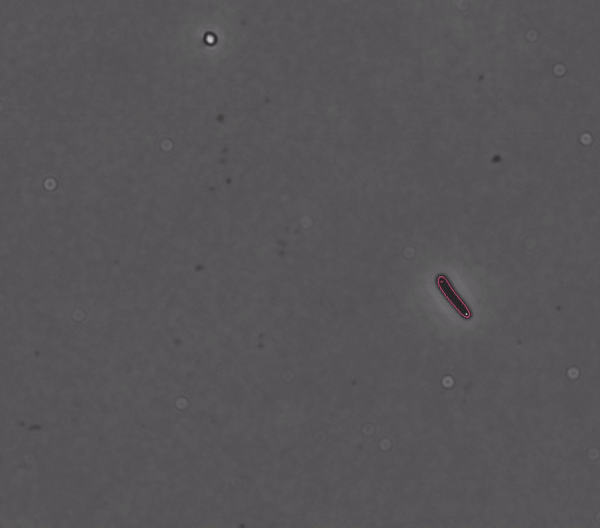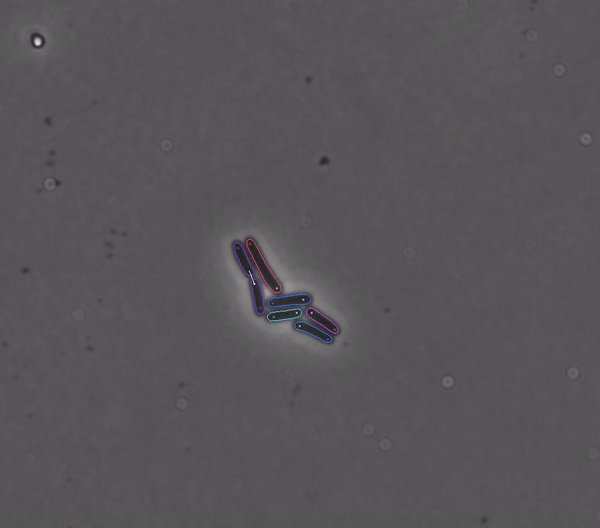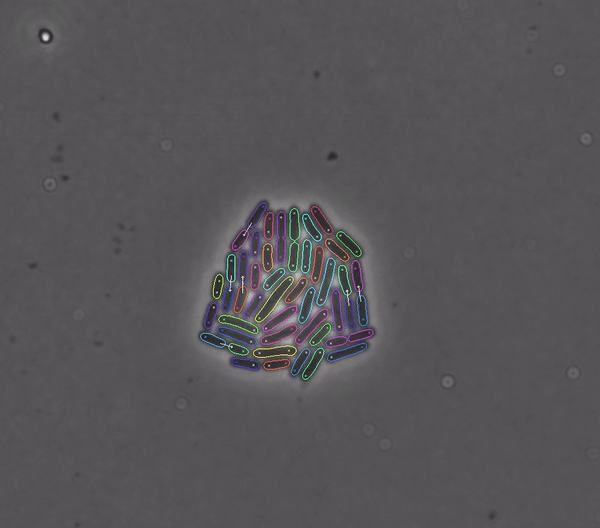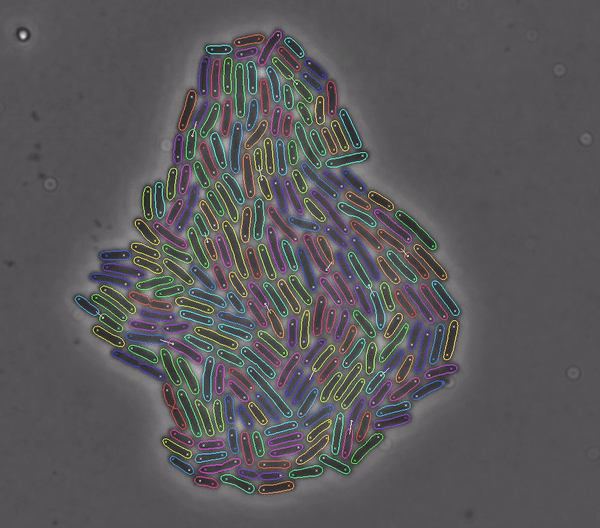
Segments and tracks bacteria
Project description
DeLTA
NOTE This is version 2 of the DeLTA pipeline. For version 1, please check out branch 'version1'
DeLTA (Deep Learning for Time-lapse Analysis) is a deep learning-based image processing pipeline for segmenting and tracking single cells in time-lapse microscopy movies.
:scroll: To get started check out the documentation at delta.readthedocs.io
:bug: If you encounter bugs or have questions about the software, please use Gitlab's issue system
For the latest hotness check out the dev branch. You can also quickly test DeLTA on our data or your own with Google Colab
for free here
See also our papers for more details:
Contributions
A big thank you to the following people who shared their data and training sets with us, they help us make DeLTA more generalizable:
- Simon van Vliet at University of Basel
- Zoran Marinković and Marianne Grognot in Philippe Nghe's group at ESPCI Paris
- Noah Olsman and Daniel Eaton in Johan Paulsson's group at Harvard
- Shuai Yang in Fan Jin's group at Shenzhen Institutes of Advanced Technology
- Jordi van Gestel and Noam Golan in Avigdor Eldar's group at Tel-Aviv University
Please reach out if you have created your own sets and think they would be helpful to the community!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file delta2-2.0.7.tar.gz.
File metadata
- Download URL: delta2-2.0.7.tar.gz
- Upload date:
- Size: 61.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.13.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
3e7c9156c80917c2fc052a0ad23117360d52001b36cbcf158de67048d9b5b20a
|
|
| MD5 |
61a3fa17d540c137047cbd94ad241bf8
|
|
| BLAKE2b-256 |
1157e7c830403951b20c8a9d25e719df200488e0de8601633947317b7a4d029c
|
File details
Details for the file delta2-2.0.7-py3-none-any.whl.
File metadata
- Download URL: delta2-2.0.7-py3-none-any.whl
- Upload date:
- Size: 56.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.13.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
40a31672c84a3c274dd660f389f30f4729cfc723a2702a15eb9b1faaa07d0f15
|
|
| MD5 |
7a713e1f1b8392ea637f22fe0c168e16
|
|
| BLAKE2b-256 |
ee1c7e06ee17a33b6ce5638046aba67b79eb402c67a279716f2ec73731bb98f9
|