Jupyter notebooks as Markdown documents, Julia, Python or R scripts
Project description
Jupyter notebooks as Markdown documents, Julia, Python or R scripts
Have you always wished Jupyter notebooks were plain text documents? Wished you could edit them in your favorite IDE? And get clear and meaningful diffs when doing version control? Then... Jupytext may well be the tool you're looking for!
Jupytext can save Jupyter notebooks as
- Markdown and R Markdown documents,
- Julia, Python, R, Bash, Scheme, Clojure, Matlab, Octave, C++ and q/kdb+ scripts.
The plain text formatting of the notebook cell divisions in scripts is configurable. Popular choices include the percent format [# %%] used by Spyder, VSCode, and others, and the light format which was developed to support this project. light uses as few cell markers as possible and is particularly suited for importing a pre-existing python script as a notebook with cell divisions automatically inferred from paragraph breaks in the source code.
There are multiple ways to use jupytext:
- Directly from Jupyter Notebook or JupyterLab. Jupytext provides a contents manager that allows Jupyter to save your notebook to your favorite format (
.py,.R,.jl,.md,.Rmd...) in addition to (or in place of) the traditional.ipynbfile. The text representation can be edited in your favorite editor. When you're done, refresh the notebook in Jupyter: inputs cells are loaded from the text file, while output cells are reloaded from the.ipynbfile if present. Refreshing preserves kernel variables, so you can resume your work in the notebook and run the modified cells without having to rerun the notebook in full. - On the command line.
jupytextconverts Jupyter notebooks to their text representation, and back. The command line tool can act on notebooks in many ways. It can synchronize multiple representations of a notebook, pipe a notebook into a reformatting tool likeblack, etc... It can also work as a pre-commit hook if you wish to automatically update the text representation when you commit the.ipynbfile. - in Vim: edit your Jupyter notebooks, represented as a Markdown document, or a Python script, with jupytext.vim.
Contents: Demo time | Example usage | Installation | Using Jupytext from within Jupyter | Scripting Jupytext | Format specifications | Want to contribute?
Demo time
Looking for a demo?
- Read the original announcement in Towards Data Science,
- Watch the PyParis talk,
- or, try Jupytext online with binder!
Example usage
Writing notebooks as plain text
You like to work with scripts? The good news is that plain scripts, which you can draft and test in your favorite IDE, open transparently as notebooks in Jupyter when using Jupytext. Run the notebook in Jupyter to generate the outputs, associate an .ipynb representation, save and share your research as either a plain script or as a traditional Jupyter notebook with outputs.
Collaborating on Jupyter Notebooks
With Jupytext, collaborating on Jupyter notebooks with Git becomes as easy as collaborating on text files.
The setup is straightforward:
- Open your favorite notebook in Jupyter notebook
- Associate a
.pyrepresentation (for instance) to that notebook - Save the notebook, and put the Python script under Git control. Sharing the
.ipynbfile is possible, but not required.
Collaborating then works as follows:
- Your collaborator pulls your script.
- The script opens as a notebook in Jupyter, with no outputs (in JupyterLab right-click the script and use the open-with context menu).
- They run the notebook and save it. Outputs are regenerated, and a local
.ipynbfile is created. - Note that, alternatively, the
.ipynbfile could have been regenerated withjupytext --sync notebook.py. - They change the notebook, and push their updated script. The diff is nothing else than a standard diff on a Python script.
- You pull the changed script, and refresh your browser. Input cells are updated. The outputs from cells that were changed are removed. Your variables are untouched, so you have the option to run only the modified cells to get the new outputs.
Code refactoring
In the animation below we propose a quick demo of Jupytext. While the example remains simple, it shows how your favorite text editor or IDE can be used to edit your Jupyter notebooks. IDEs are more convenient than Jupyter for navigating through code, editing and executing cells or fractions of cells, and debugging.
- We start with a Jupyter notebook.
- The notebook includes a plot of the world population. The plot legend is not in order of decreasing population, we'll fix this.
- We want the notebook to be saved as both a
.ipynband a.pyfile: we select Pair Notebook with a light Script in either the Jupytext menu in Jupyter Notebook, or in the Jupytext commands in JupyterLab. This has the effect of adding a"jupytext": {"formats": "ipynb,py:light"},entry to the notebook metadata. - The Python script can be opened with PyCharm:
- Navigating in the code and documentation is easier than in Jupyter.
- The console is convenient for quick tests. We don't need to create cells for this.
- We find out that the columns of the data frame were not in the correct order. We update the corresponding cell, and get the correct plot.
- The Jupyter notebook is refreshed in the browser. Modified inputs are loaded from the Python script. Outputs and variables are preserved. We finally rerun the code and get the correct plot.
Importing Jupyter Notebooks as modules
Jupytext allows to import code from other Jupyter notebooks in a very simple manner. Indeed, all you need to do is to pair the notebook that you wish to import with a script, and import the resulting script.
If the notebook contains demos and plots that you don't want to import, mark those cells as either
- active only in the
ipynbformat, with the{"active": "ipynb"}cell metadata, or with anactive-ipynbtag (you may use the jupyterlab-celltags extension for this). - frozen, using the freeze extension for Jupyter notebook.
Inactive cells will be commented in the paired script, and consequently will not be executed when the script is imported.
Installation
Jupytext is available on pypi and on conda-forge. Run either of
pip install jupytext --upgrade
or
conda install -c conda-forge jupytext
If you want to use Jupytext within Jupyter Notebook or JupyterLab, make sure you install Jupytext in the Python environment where the Jupyter server runs. If that environment is read-only, for instance if your server is started using JupyterHub, install Jupytext in user mode with:
/path_to_your_jupyter_environment/python -m pip install jupytext --upgrade --user
Jupytext's contents manager
Jupytext includes a contents manager for Jupyter that allows Jupyter to open and save notebooks as text files. When Jupytext's content manager is active in Jupyter, scripts and Markdown documents have a notebook icon.
If you don't have the notebook icon on text documents after a fresh restart of your Jupyter server, install the contents manager manually. Append
c.NotebookApp.contents_manager_class = "jupytext.TextFileContentsManager"
to your .jupyter/jupyter_notebook_config.py file (generate a Jupyter config, if you don't have one yet, with jupyter notebook --generate-config). Our contents manager accepts a few options: default formats, default metadata filter, etc — read more on this below. Then, restart Jupyter Notebook or JupyterLab, either from the JupyterHub interface or from the command line with
jupyter notebook # or lab
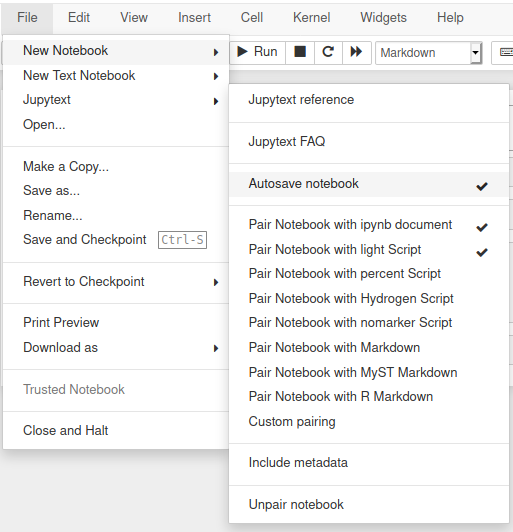
Jupytext menu in Jupyter Notebook
Jupytext includes an extensions for Jupyter Notebook that adds a Jupytext section in the File menu.
If the extension was not automatically installed, install and activate it with
jupyter nbextension install --py jupytext [--user]
jupyter nbextension enable --py jupytext [--user]
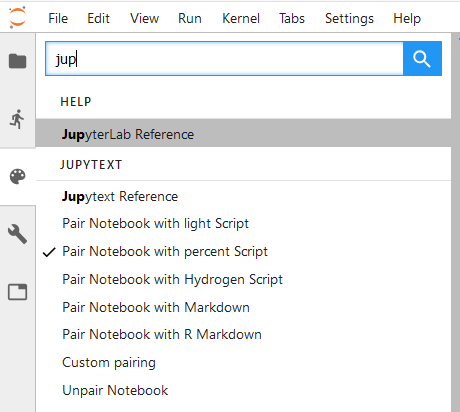
Jupytext commands in JupyterLab
In JupyterLab, Jupytext adds a set of commands to the command palette:
If you don't see these commands, install the extension manually with
jupyter labextension install jupyterlab-jupytext
(the above requires npm, run conda install nodejs first if you don't have npm).
Using Jupytext from within Jupyter
Paired notebooks
Jupytext can write a given notebook to multiple files. In addition to the original notebook file, Jupytext can save the input cells to a text file — either a script or a Markdown document. Put the text file under version control for a clear commit history. Or refactor the paired script, and reimport the updated input cells by simply refreshing the notebook in Jupyter.
Per-notebook configuration
Select the pairing for a given notebook using either the Jupytext menu in Jupyter Notebook, or the Jupytext commands in JupyterLab.
These command simply add a "jupytext": {"formats": "ipynb,md"}-like entry in the notebook metadata. You could also set that metadata yourself with Edit/Edit Notebook Metadata in Jupyter Notebook. In JupyterLab, use this extension.
The pairing information for one or multiple notebooks can be set on the command line:
jupytext --set-formats ipynb,py [--sync] notebook.ipynb
You can pair a notebook to as many text representations as you want (see our World population notebook in the demo folder). Format specifications are of the form
[[path/][prefix]/][suffix.]ext[:format_name]
where
extis one ofipynb,md,Rmd,jl,py,R,sh,cpp,q. Use theautoextension to have the script extension chosen according to the Jupyter kernel.format_name(optional) is eitherlight(default for scripts),bare,percent,hydrogen,sphinx(Python only),spin(R only) — see below for the format specifications.path,prefixandsuffixallow to save the text representation to files with different names, or in a different folder.
If you want to pair a notebook to a python script in a subfolder named scripts, set the formats metadata to ipynb,scripts//py. If the notebook is in a notebooks folder and you want the text representation to be in a scripts folder at the same level, set the Jupytext formats to notebooks//ipynb,scripts//py.
Jupytext accepts a few additional options. These options should be added to the "jupytext" section in the metadata — use either the metadata editor or the --opt/--format-options argument on the command line.
comment_magics: By default, Jupyter magics are commented when notebooks are exported to any other format than markdown. If you prefer otherwise, use this boolean option, or is global counterpart (see below).notebook_metadata_filter: By default, Jupytext only exports thekernelspecandjupytextmetadata to the text files. Set"jupytext": {"notebook_metadata_filter": "-all"}if you want that the script has no notebook metadata at all. The value fornotebook_metadata_filteris a comma separated list of additional/excluded (negated) entries, withalla keyword that allows to exclude all entries.cell_metadata_filter: By default, cell metadataautoscroll,collapsed,scrolled,trustedandExecuteTimeare not included in the text representation. Add or exclude more cell metadata with this option.
Global configuration
Jupytext's contents manager also accepts global options. These options have to be set on Jupytext's contents manager, so please first include the following line in your .jupyter/jupyter_notebook_config.py file:
c.NotebookApp.contents_manager_class = "jupytext.TextFileContentsManager"
We start with the default format pairing. Say you want to always associate every .ipynb notebook with a .md file (and reciprocally). This is simply done by adding the following to your Jupyter configuration file:
# Always pair ipynb notebooks to md files
c.ContentsManager.default_jupytext_formats = "ipynb,md"
(and similarly for the other formats).
In case the percent format is your favorite, add the following to your .jupyter/jupyter_notebook_config.py file:
# Use the percent format when saving as py
c.ContentsManager.preferred_jupytext_formats_save = "py:percent"
and then, Jupytext will understand "jupytext": {"formats": "ipynb,py"} as an instruction to create the paired Python script in the percent format.
To disable global pairing for an individual notebook, set formats to a single format, e.g.:
"jupytext": {"formats": "ipynb"}
Metadata filtering
You can specify which metadata to include or exclude in the text files created by Jupytext by setting c.ContentsManager.default_notebook_metadata_filter (notebook metadata) and c.ContentsManager.default_cell_metadata_filter (cell metadata). They accept a string of comma separated keywords. A minus sign - in font of a keyword means exclusion.
Suppose you want to keep all the notebook metadata but widgets and varInspector in the YAML header. For cell metadata, you want to allow ExecuteTime and autoscroll, but not hide_output. You can set
c.ContentsManager.default_notebook_metadata_filter = "all,-widgets,-varInspector"
c.ContentsManager.default_cell_metadata_filter = "ExecuteTime,autoscroll,-hide_output"
If you want that the text files created by Jupytext have no metadata, you may use the global metadata filters below. Please note that with this setting, the metadata is only preserved in the .ipynb file.
c.ContentsManager.default_notebook_metadata_filter = "-all"
c.ContentsManager.default_cell_metadata_filter = "-all"
NB: All these global options (and more) are documented here.
Can I edit a notebook simultaneously in Jupyter and in a text editor?
When saving a paired notebook using Jupytext's contents manager, Jupyter updates both the .ipynb and its text representation. The text representation can be edited outside of Jupyter. When the notebook is refreshed in Jupyter, the input cells are read from the text file, and the output cells from the .ipynb file.
It is possible (and convenient) to leave the notebook open in Jupyter while you edit its text representation. However, you don't want that the two editors save the notebook simultaneously. To avoid this:
- deactivate Jupyter's autosave, by toggling the
"Autosave notebook"menu entry (or run%autosave 0in a cell of the notebook) - and refresh the notebook when you switch back from the editor to Jupyter.
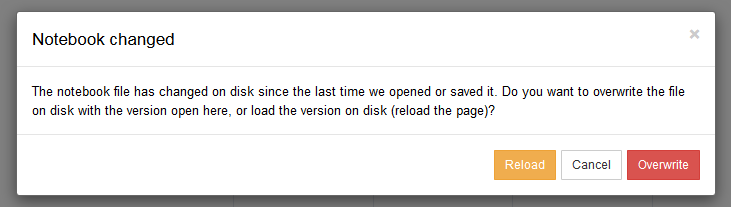
In case you forgot to refresh, and saved the Jupyter notebook while the text representation had changed, no worries: Jupyter will ask you which version you want to keep:
When that occurs, please choose the version in which you made the latest changes. And give a second look to our advice to deactivate the autosaving of notebooks in Jupyter.
How to open scripts with either the text or notebook view in Jupyter?
With Jupytext's contents manager for Jupyter, scripts and Markdown documents gain a notebook icon. If you don't see the notebook icon, double check the contents manager configuration.
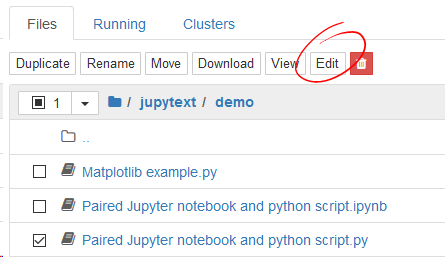
By default, Jupyter Notebook open scripts and Markdown documents as notebooks. If you want to open them with the text editor, select the document and click on edit:
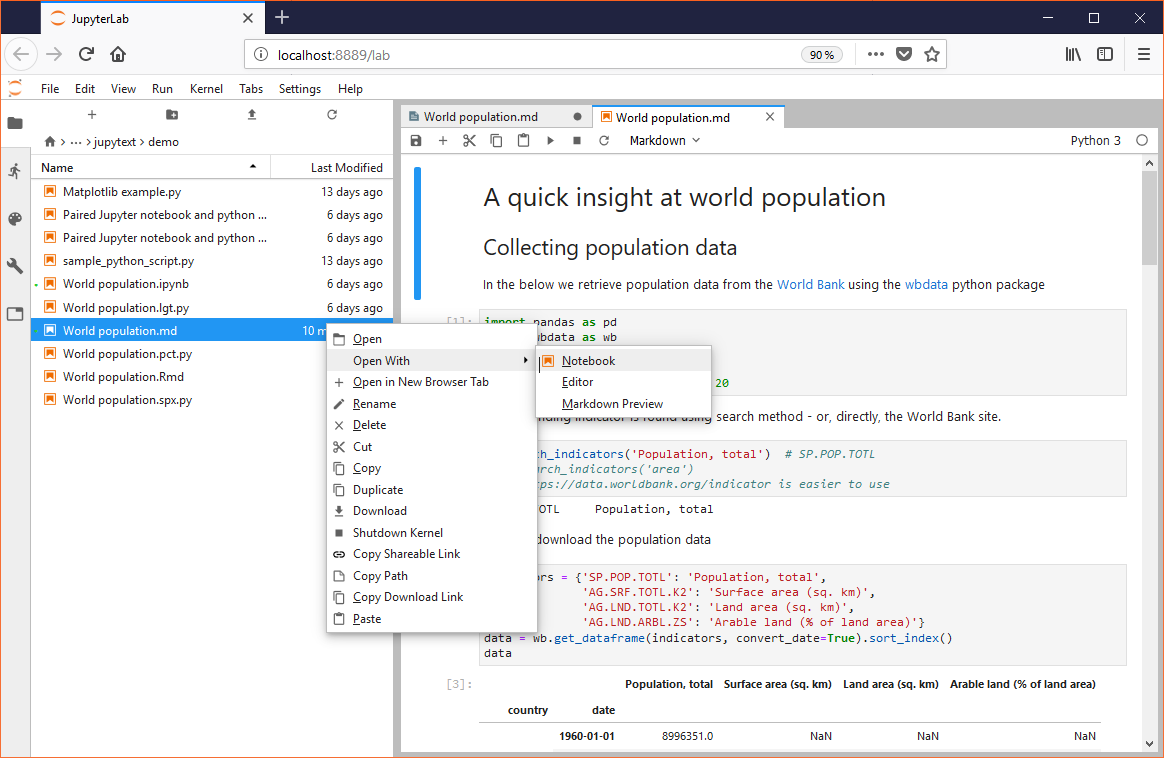
In JupyterLab this is slightly different. Scripts and Markdown document also have a notebook icon. But they open as text by default. Open them as notebooks with the Open With -> Notebook context menu (available in JupyterLab 0.35 and above):
If do not want to classify scripts or Markdown documents as notebooks, please use the notebook_extension option. For instance, if you want to get the notebook icon only for .ipynb and .Rmd files, set
c.ContentsManager.notebook_extensions = "ipynb,Rmd"
Please note that, with the above setting, Jupyter will not let you open scripts as notebooks. If you still want to do so, use Jupytext command line (see below) to first convert or pair the script to an .ipynb notebook.
Scripting Jupytext
Command line conversion
The package provides a jupytext script for command line conversion between the various notebook extensions:
jupytext --to py notebook.ipynb # create a notebook.py file in the light format
jupytext --to py:percent notebook.ipynb # create a notebook.py file in the double percent format
jupytext --to py:percent --comment-magics false notebook.ipynb # create a notebook.py file in the double percent format, and do not comment magic commands
jupytext --to markdown notebook.ipynb # create a notebook.md file
jupytext --output script.py notebook.ipynb # create a script.py file
jupytext --to notebook notebook.py # overwrite notebook.ipynb (remove outputs)
jupytext --to notebook --update notebook.py # update notebook.ipynb (preserve outputs)
jupytext --to ipynb notebook1.md notebook2.py # overwrite notebook1.ipynb and notebook2.ipynb
jupytext --to md --test notebook.ipynb # Test round trip conversion
jupytext --to md --output - notebook.ipynb # display the markdown version on screen
jupytext --from ipynb --to py:percent # read ipynb from stdin and write double percent script on stdout
Jupytext has a --sync mode that updates all the paired representations of a notebook based on the file that was last modified. You may also find useful to --pipe the text representation of a notebook into tools like black:
jupytext --sync --pipe black notebook.ipynb # read most recent version of notebook, reformat with black, save
The jupytext command accepts many arguments. Use the --set-formats and the --update-metadata arguments to edit the pairing information or more generally the notebook metadata. Execute jupytext --help to access the documentation.
Jupytext as a Git pre-commit hook
Jupytext is also available as a Git pre-commit hook. Use this if you want Jupytext to create and update the .py (or .md...) representation of the staged .ipynb notebooks. All you need is to create an executable .git/hooks/pre-commit file with the following content:
#!/bin/sh
# For every ipynb file in the git index, add a Python representation
jupytext --from ipynb --to py:light --pre-commit
#!/bin/sh
# For every ipynb file in the git index:
# - apply black and flake8
# - export the notebook to a Python script in folder 'python'
# - and add it to the git index
jupytext --from ipynb --pipe black --check flake8 --pre-commit
jupytext --from ipynb --to python//py:light --pre-commit
If you don't want notebooks to be committed (and only commit the representations), you can ask the pre-commit hook to unstage notebooks after conversion by adding the following line:
git reset HEAD **/*.ipynb
Note that these hooks do not update the .ipynb notebook when you pull. Make sure to either run jupytext in the other direction, or to use our paired notebook and our contents manager for Jupyter. Also, Jupytext does not offer a merge driver. If a conflict occurs, solve it on the text representation and then update or recreate the .ipynb notebook. Or give a try to nbdime and its merge driver.
Testing the round-trip conversion
Representing Jupyter notebooks as scripts requires a solid round trip conversion. You don't want your notebooks (nor your scripts) to be modified because you are converting them to the other form. Our test suite includes a few hundred tests to ensure that round trip conversion is safe.
You can easily test that the round trip conversion preserves your Jupyter notebooks and scripts. Run for instance:
# Test the ipynb -> py:percent -> ipynb round trip conversion
jupytext --test notebook.ipynb --to py:percent
# Test the ipynb -> (py:percent + ipynb) -> ipynb (à la paired notebook) conversion
jupytext --test --update notebook.ipynb --to py:percent
Note that jupytext --test compares the resulting notebooks according to its expectations. If you wish to proceed to a strict comparison of the two notebooks, use jupytext --test-strict, and use the flag -x to report with more details on the first difference, if any.
Please note that
- Scripts opened with Jupyter have a default metadata filter that prevents additional notebook or cell metadata to be added back to the script. Remove the filter if you want to store Jupytext's settings, or the kernel information, in the text file.
- Cell metadata are available in the
lightandpercentformats, as well as in the Markdown and R Markdown formats. R scripts inspinformat support cell metadata for code cells only. Sphinx Gallery scripts insphinxformat do not support cell metadata. - By default, a few cell metadata are not included in the text representation of the notebook. And only the most standard notebook metadata are exported. Learn more on this in the sections for notebook specific and global settings for metadata filtering.
Reading notebooks in Python
You can also manipulate notebooks in a Python shell or script using Jupytext's main functions:
# Read a notebook from a file. Format can be any of 'py', 'md', 'jl:percent', ...
readf(nb_file, fmt=None)
# Read a notebook from a string. Here, format should contain at least the file extension.
reads(text, fmt)
# Return the text representation for a notebook in the desired format.
writes(notebook, fmt)
# Write a notebook to a file in the desired format.
writef(notebook, nb_file, fmt=None)
Format specifications
Markdown and R Markdown
Save Jupyter notebooks as Markdown documents and edit them in one of the many editors with good Markdown support.
R Markdown is RStudio's format for notebooks, with support for R, Python, and many other languages.
Jupytext's implementation for Jupyter notebooks as Markdown or R Markdown documents is as follows:
- The notebook metadata (Jupyter kernel, etc) goes to a YAML header
- Code and raw cells are encoded as Markdown code blocks with triple backticks. In a Python notebook, a code cell starts with
```pythonand ends with```. Cell metadata are found after the language information, with akey=valuesyntax, wherevalueis encoded in JSON format (Markdown) or R format (R Markdown). R Markdown code cell options are mapped to the corresponding Jupyter cell metadata options, when available. - Markdown cells are inserted verbatim and separated with two blank lines. When required (cells with metadata, cells that contain two blank lines or code blocks), Jupytext protects the cell boundary with HTML comments:
<!-- #region -->and<!-- #endregion -->. Cells with explicit boundaries are foldable in vscode, and can accept both a title and/or metadata in JSON format:<!-- #region This is the title for my protected cell {"key": "value"}-->.
See how our World population.ipynb notebook in the demo folder is represented in Markdown or R Markdown.
When you open a plain Markdown file in Jupytext, the Markdown text is rendered in Markdown cells. Cells breaks occur when the text contains two consecutive blank lines (or code cells). If you want to also split cells on Markdown headers, so that headers prefixed by one blank line appear at the top of a new cell, use the split_at_heading option. Set the option either on the command line, or by adding "split_at_heading": true to the jupytext section in the notebook metadata, or on Jupytext's content manager:
c.ContentsManager.split_at_heading = True
Pandoc's Markdown
Pandoc, the Universal document converter, can now read and write Jupyter notebooks - see Pandoc's documentation.
Pandoc's Markdown format is available in Jupytext as md:pandoc. This requires pandoc in version 2.7.2 or above - please check Pandoc's version number with pandoc -v before running either jupytext command line, or before you start your Jupyter notebook or Lab server. Note that you can get the latest version of pandoc in a conda environment with
conda install pandoc -c conda-forge
Pandoc's format uses Pandoc divs (:::) as explicit cell markers. See how our World population.ipynb notebook is represented in that format. Please also note that pandoc, while preserving the HTML rendering, may reformat the text in some of the Markdown cells. If that is an issue for you, please wait until jgm/pandoc#5408 gets implemented.
Jupytext currently strips the output cells before calling pandoc. As for the other formats, outputs cells can be preserved in paired notebooks. In the case of the pandoc format, paired notebooks are available with the metadata "jupytext": {"formats": "md:pandoc,ipynb"}.
The light format for notebooks as scripts
The light format was created for this project. It is the default format for scripts. That format can read any script as a Jupyter notebook, even scripts which were never prepared to become a notebook. When a notebook is written as a script using this format, only a few cells markers are introduced—none if possible.
The light format has:
- A (commented) YAML header, that contains the notebook metadata.
- Markdown cells are commented, and separated from other cells with a blank line.
- Code cells are exported verbatim (except for Jupyter magics, which are commented), and separated with blank lines. Code cells are reconstructed from consistent Python paragraphs (no function, class or multiline comment will be broken).
- Cells that contain more than one Python paragraphs need an explicit start-of-cell delimiter that is, by default,
# +(// +in C++, etc). Cells that have explicit metadata have a cell header# + {JSON}where the metadata is represented, in JSON format. The default end of cell delimiter is# -, and is omitted when followed by another explicit start of cell marker.
The light format is currently available for Python, Julia, R, Bash, Scheme, Clojure, Matlab, Octave, C++, q/kdb+ and IDL. Open our sample notebook in the light format here.
A variation of the light format is the bare format, with no cell marker at all. Please note that this format will split your code cells on code paragraphs. By default, this format still includes a YAML header - if you prefer to also remove the header, set "notebook_metadata_filter": "-all" in the jupytext section of your notebook metadata.
The light format can use custom cell markers instead of # + or # -. If you prefer to mark cells with VScode/PyCharm (resp. Vim) folding markers, set "cell_markers": "region,endregion" (resp. "{{{,}}}") in the jupytext section of the notebook metadata. If you want to configure this as a global default, add either
c.ContentsManager.default_cell_markers = "region,endregion" # Use VScode/PyCharm region folding delimiters
or
c.ContentsManager.default_cell_markers = "{{{,}}}" # Use Vim region folding delimiters
to your .jupyter/jupyter_notebook_config.py file.
The percent format
The percent format is a representation of Jupyter notebooks as scripts, in which cells are delimited with a commented double percent sign # %%. The format was introduced by Spyder five years ago, and is now supported by many editors, including
- Spyder IDE,
- Hydrogen, a package for Atom,
- VS Code with the vscodeJupyter extension,
- Python Tools for Visual Studio,
- and PyCharm Professional.
Our implementation of the percent format is compatible with the original specifications by Spyder. We extended the format to allow markdown cells and cell metadata. Cell headers have the following structure:
# %% Optional text [cell type] {optional JSON metadata}
where cell type is either omitted (code cells), or [markdown] or [raw]. The content of markdown and raw cells is commented out in the resulting script.
Percent scripts created by Jupytext have a header with an explicit format information. The format of scripts with no header is inferred automatically: scripts with at least one # %% cell are identified as percent scripts. Scripts with at least one double percent cell, and an uncommented Jupyter magic command, are identified as hydrogen scripts.
The percent format is currently available for Python, Julia, R, Bash, Scheme, Clojure, Matlab, Octave, C++, q/kdb+ and IDL. Open our sample notebook in the percent format here.
If the percent format is your favorite one, add the following to your .jupyter/jupyter_notebook_config.py file:
c.ContentsManager.preferred_jupytext_formats_save = "py:percent" # or "auto:percent"
Then, Jupytext's content manager will understand "jupytext": {"formats": "ipynb,py"}, as an instruction to create the paired Python script in the percent format.
By default, Jupyter magics are commented in the percent representation. If you run the percent scripts in Hydrogen, use instead the hydrogen format, a variant of the percent format that does not comment Jupyter magic commands.
Sphinx-gallery scripts
Another popular notebook-like format for Python scripts is the Sphinx-gallery format. Scripts that contain at least two lines with more than twenty hash signs are classified as Sphinx-Gallery notebooks by Jupytext.
Comments in Sphinx-Gallery scripts are formatted using reStructuredText rather than markdown. They can be converted to markdown for a nicer display in Jupyter by adding a c.ContentsManager.sphinx_convert_rst2md = True line to your Jupyter configuration file. Please note that this is a non-reversible transformation—use this only with Binder. Revert to the default value sphinx_convert_rst2md = False when you edit Sphinx-Gallery files with Jupytext.
Turn a GitHub repository containing Sphinx-Gallery scripts into a live notebook repository with Binder and Jupytext by adding only two files to the repo:
binder/requirements.txt, a list of the required packages (includingjupytext).jupyter/jupyter_notebook_config.pywith the following contents:
c.NotebookApp.contents_manager_class = "jupytext.TextFileContentsManager"
c.ContentsManager.preferred_jupytext_formats_read = "py:sphinx"
c.ContentsManager.sphinx_convert_rst2md = True
Our sample notebook is also represented in sphinx format here.
R knitr::spin scripts
The spin format is specific to R scripts. These scripts can be compiled into reports using either knitr::spin or RStudio. The implementation of the format is as follows:
- Jupyter metadata are in YAML format, in a
#'-commented header. - Markdown cells are commented with
#'. - Code cells are exported verbatim. Cell metadata are signalled with
#+. Cells end with a blank line, an explicit start of cell marker, or a markdown cell.
Fine tuning
Jupyter magic commands are commented when exporting the notebook to text, except for the markdown and the hydrogen format. If you want to change this for a single line, add a #escape or #noescape flag on the same line as the magic, or a "comment_magics": true or false entry in the notebook metadata, in the "jupytext" section. Or set your preference globally on the contents manager by adding this line to .jupyter/jupyter_notebook_config.py:
c.ContentsManager.comment_magics = True # or False
Also, you may want some cells to be active only in the Python, or R Markdown representation. For this, use the active cell metadata. Set "active": "ipynb" if you want that cell to be active only in the Jupyter notebook. And "active": "py" if you want it to be active only in the Python script. And "active": "ipynb,py" if you want it to be active in both, but not in the R Markdown representation...
Extending the light and percent formats to more languages
You want to extend the light and percent format to another language? In principle that is easy, and you will only have to:
- document the language extension and comment by adding one line to
_SCRIPT_EXTENSIONSinlanguages.py. - contribute a sample notebook in
tests\notebooks\ipynb_[language]. - add two tests in
test_mirror.py: one for thelightformat, and another one for thepercentformat. - Make sure that the tests pass, and that the text representations of your notebook, found in
tests\notebooks\mirror\ipynb_to_scriptandtests\notebooks\mirror\ipynb_to_percent, are valid scripts.
Want to contribute?
Contributions are welcome. Please let us know how you use jupytext and how we could improve it. You think the documentation could be improved? Go ahead! And stay tuned for more demos on medium and twitter!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file jupytext-1.1.3.tar.gz.
File metadata
- Download URL: jupytext-1.1.3.tar.gz
- Upload date:
- Size: 139.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.5.0.1 requests/2.21.0 setuptools/41.0.0 requests-toolbelt/0.9.1 tqdm/4.32.1 CPython/3.7.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
d09f8099394d3b96f604f2b64287a6bb4b9cd701bf6aa5d62bebdd39c6e88cce
|
|
| MD5 |
2fc125d1c35d45bffa775c2ded10b675
|
|
| BLAKE2b-256 |
986d2c3b61c1fb281d4199748294777074ad6805cf070de0f24e46ca90c95881
|