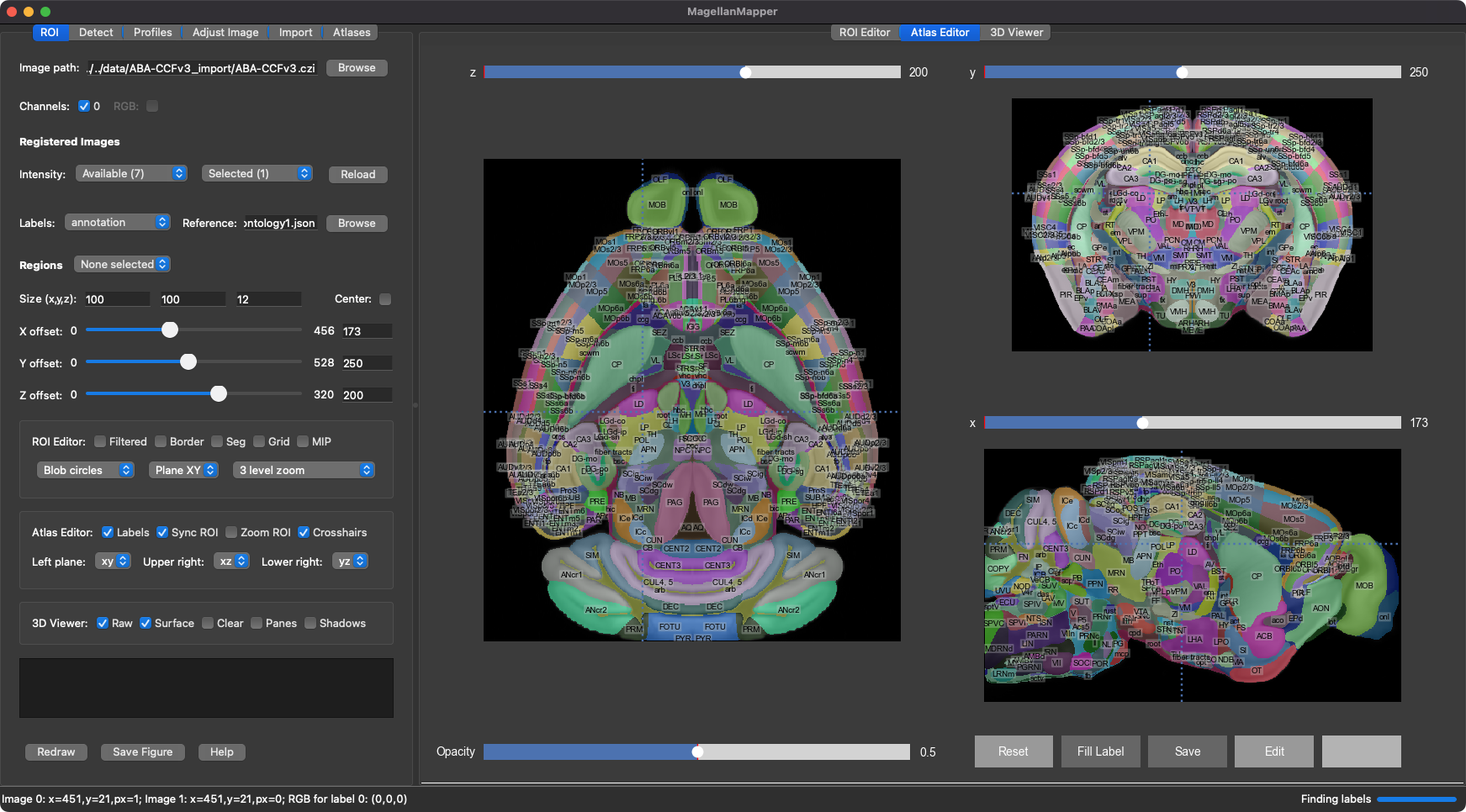
3D atlas analysis and annotation
Project description
MagellanMapper
MagellanMapper is a graphical imaging informatics suite for 3D reconstruction and automated analysis of whole specimens and atlases. Its design philosophy is to make the raw 3D images as accessible as possible, simplify annotation from nuclei to atlases, and scale from the laptop or desktop to the cloud in cross-platform environments.
Quick Reference
- NEW: Docs are now on ReadTheDocs!
- Installation (more details)
- Intro to running MagellanMapper
- Using the viewers
- Command-line interface
- Configuration and settings
- Publications
Installation
If you use Conda (available here), you can install MagellanMapper into a new environment:
conda env create -n mag -f https://raw.githubusercontent.com/sanderslab/magellanmapper/master/envs/environment_rel.yml
To run in this new environment named mag*:
conda activate mag
mm
* mm was added in v1.6.0 to launch from installed packages
Or install using Pip (virtual environment recommended):
pip install "magellanmapper[most]" --extra-index-url https://pypi.fury.io/dd8/
The extra index accesses a few customized dependencies for MagellanMapper.
Conda installs Java to import proprietary image file formats, which can also be installed separately when using Pip (eg from here).
For the latest updates and fixes, download from Git and install:
git clone https://github.com/sanderslab/magellanmapper.git
conda env create -n mag -f environment.yml
pip install -e ".[most]" --extra-index-url https://pypi.fury.io/dd8/
python run.py
Or for Pip, replace "magellanmapper[most]" with ".[most]".
More ways to install and run
See the install docs for more details, including:
- Installer scripts, which install Miniconda and MagellanMapper without requiring command-line usage
- Installer packages, for point-and-click installation
Using MagellanMapper
MagellanMapper consists of a graphical user interface (GUI), command-line interface (CLI), and application programming interface (API) for Python programmatic access. See the GUI docs for graphical usage and the CLI docs for scripting.
For automated tasks, see the sample_cmds_bash.ipynb Jupyter Notebook (or the older sample_cmds.sh script) that shows examples of running the CLI and exploring images in the GUI. See ReadTheDocs for more details, including viewer shortcuts and customizing settings for your image analysis.
Have a question? Found a bug? Want a feature? Please ask!
Image file import
Large images or proprietary microscopy formats such as CZI can be imported by MagellanMapper into NumPy format, which allows on-the-fly loading to reduce memory requirements and initial loading time. In the "Import" tab, you can select files, view and update metadata, and import the files.
Medical imaging formats such as .mha (or .mhd/.raw) and .nii (or .nii.gz) can be opened with the SimpleITK/SimpleElastix library and do not require separate import.
Sample 3D data
To try out functions with sample images, download any of these files:
- Sample region of nuclei at 4x (
sample_region.zip) - Sample downsampled tissue cleared whole brain (
sample_brain.zip) - Allen Developing Mouse Brain Atlas E18.5 (
ADMBA-E18pt5.zip)
Related publications and datasets
- For more information on the methods used for 3D atlas construction, please see: https://elifesciences.org/articles/61408
- For step-by-step instructions on using v1.3.x of the software, please see: https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/cpns.104 (now open access on PubMed!); see ReadTheDocs for ongoing updates
- The 3D reconstructed versions of the Allen Developing Mouse Brain Atlas: https://search.kg.ebrains.eu/instances/Project/b8a8e2d3-4787-45f2-b010-589948c33f20
- Sample wild-type whole mouse brains at age P0: https://search.kg.ebrains.eu/instances/Dataset/2423e103-35e9-40cf-ab0c-0e3d08d24d5a
Licensed under the open-source BSD-3 license
Author: David Young, 2017, 2023, Stephan Sanders Lab
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file magellanmapper-1.6b3.tar.gz.
File metadata
- Download URL: magellanmapper-1.6b3.tar.gz
- Upload date:
- Size: 449.1 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
56b8fabba2232bc11fa436ba734c60cdd09cbacd4f3702dc0a503be6e81d31e4
|
|
| MD5 |
2d688e7ccf59d06e86d711efd9f4aae9
|
|
| BLAKE2b-256 |
ecb229d49a6e2594a9eb5db2ca58f0a0868b45e47186c5a4f5478deaf13aa1ed
|
File details
Details for the file magellanmapper-1.6b3-py3-none-any.whl.
File metadata
- Download URL: magellanmapper-1.6b3-py3-none-any.whl
- Upload date:
- Size: 485.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c86aebedce3a32d247807e3a078a155e3eee4365c20c844a7efe3f8c5dfb2a5f
|
|
| MD5 |
57f5ab2b2e32e689bc6f0dfdb41c358c
|
|
| BLAKE2b-256 |
09af83663c09eaf24ff52c653a7e0bdeabeeeadef0a1dde5756ffeaca2b51856
|