MESSI: Multi Ensamble Strategy for Structural Elucidation
Reason this release was yanked:
fail
Project description
MESSI :atom:
Multi Ensamble Strategy for Structural identification
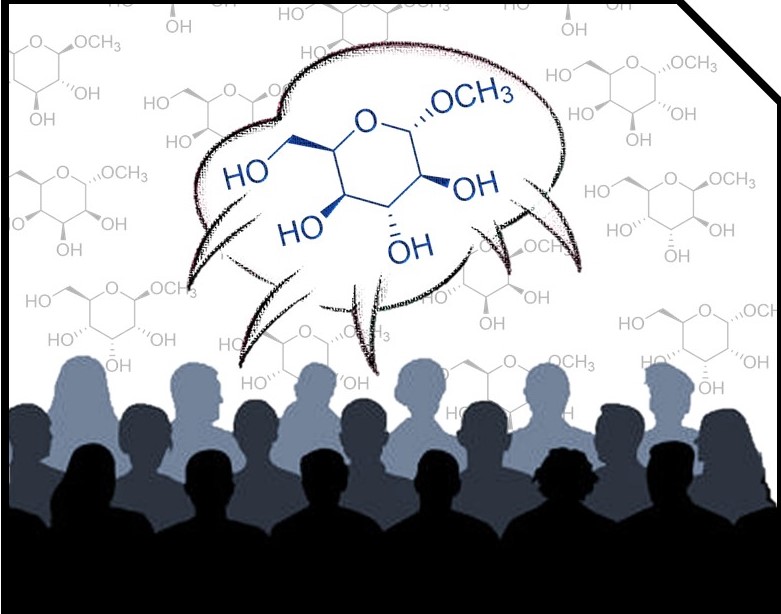
This repository contains all codes and data required to run MESSI calculations.
Description
MESSI is a Python program to compute Multi Ensamble DP4+ probabilities in the stereochemical assignament of new organic compounds.
Installation Requirements
MESSI.py needs python 3.8 or later to work. The module can be installed by console using:
pip3 install messi_nmr
Usage by console: messi
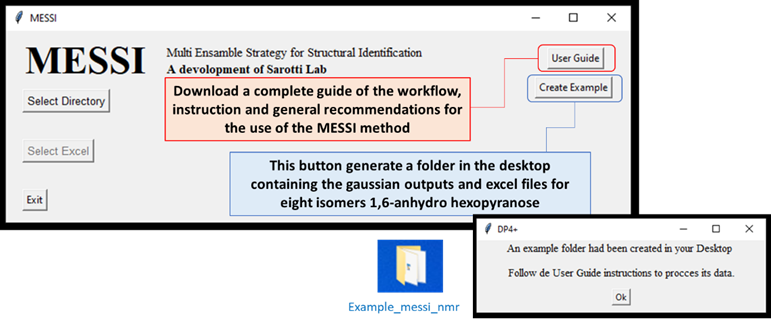
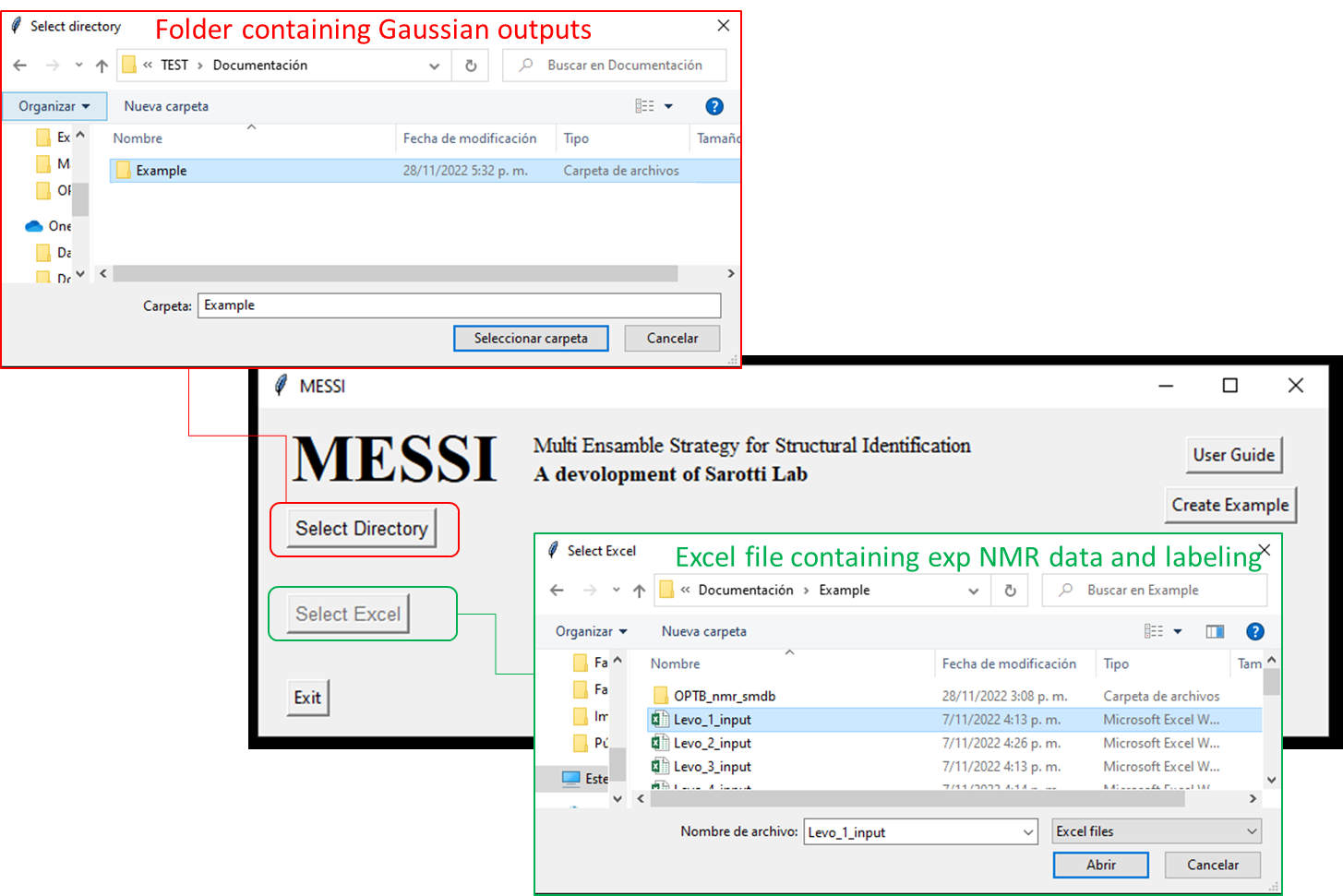
Installing the python module will automatically generate a
messi.pyshortcut on your desktop, which allows direct execution of the program without the use of a console. In order to test the correct software operation is recommended to run the provided example, which could be download by clicking the buttomCreate Example. This will create a folder nameExample_messi_nmrin desktop containing all the files needed by de use of MESSI.
User Guide
Terms of use. You need to create a folder containing the following files:
1) The outputs of the NMR and SCF calculations (all conformers for all isomers).
2) An excel file containing the experimental data and the labels of each nucleus associated with each experimental value.
1) The output files: must be named following the next convention: n_*_m*.log or .out, where n identifies the ith isomer, ranging from 1 to N where N is the number of candidate structures under study, and m indicate the conformer number. For instance:
1_NewNatProd_c01.log (Conformer 1 of isomer 1 of a compound named NewNatProd)
1_NewNatProd_c02.log (Conformer 2 of isomer 1 of a compound named NewNatProd)
2_NewNatProd_c01.log (Conformer 1 of isomer 2 of a compound named NewNatProd)
2_NewNatProd_c02.log (Conformer 2 of isomer 2 of a compound named NewNatProd)
The NMR and SCRF/SMD energies calculation could be in the same or different outputs. If they are separated, both file must begin with the same name (
n_*_m*) and a suffix must be added in order to differentiate the calculation type. If the number of files for NMR calculations does not match the number of SMD energy calculations the script will not run.
The script handles outputs from Gaussian 09 and 16.
Note: Name files adequately is essential in order to match SMD energy with the NMR data for each conformer.
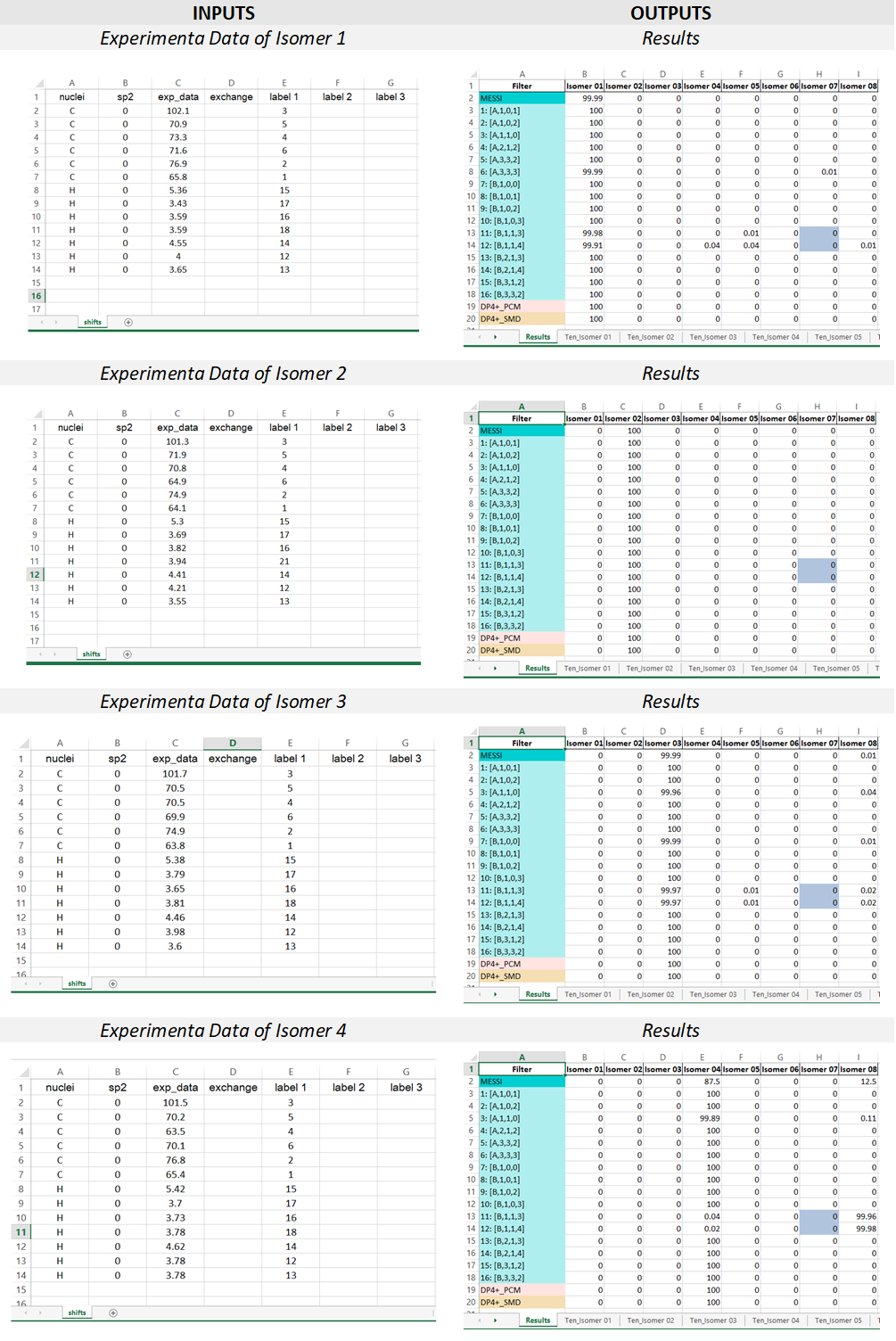
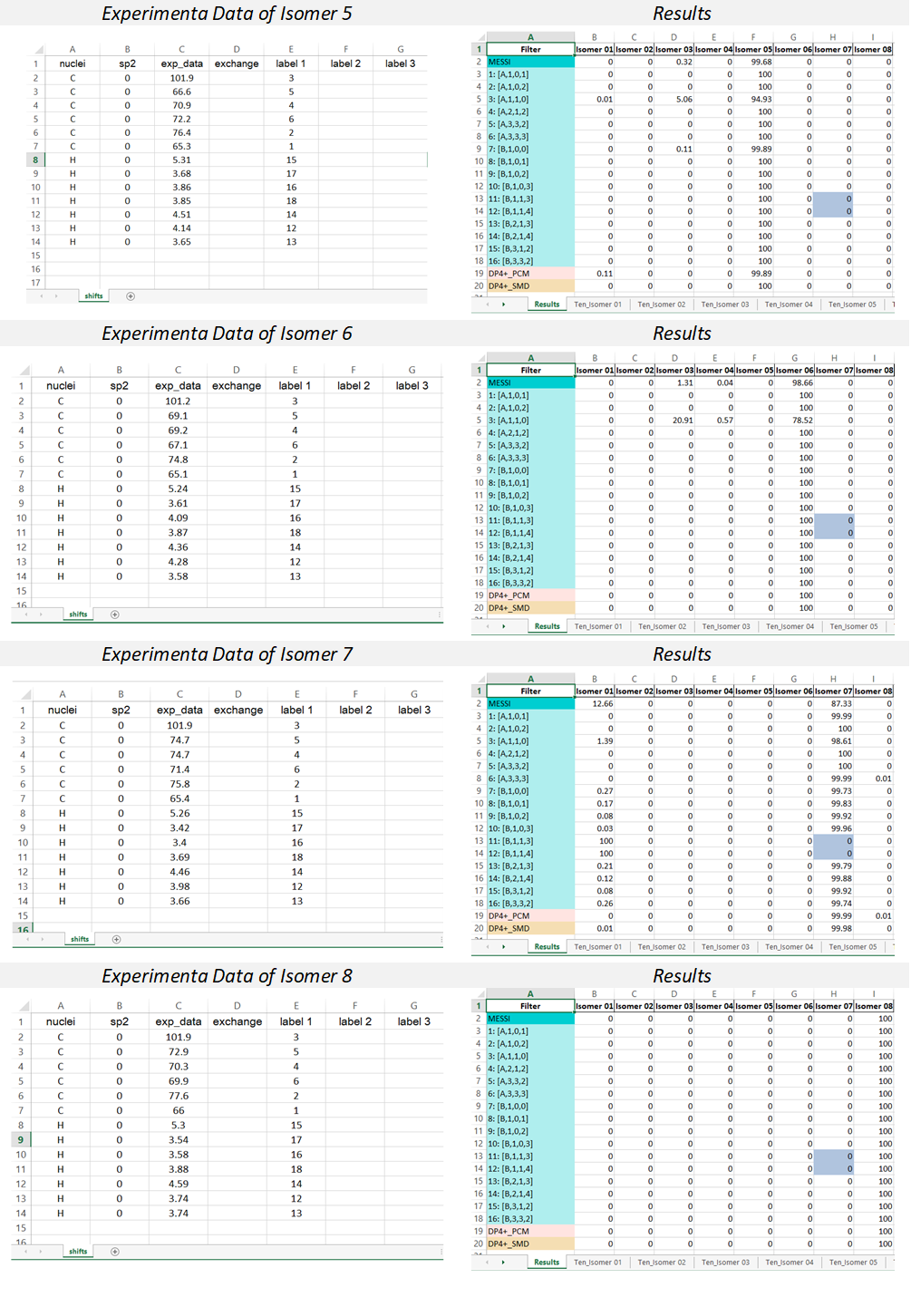
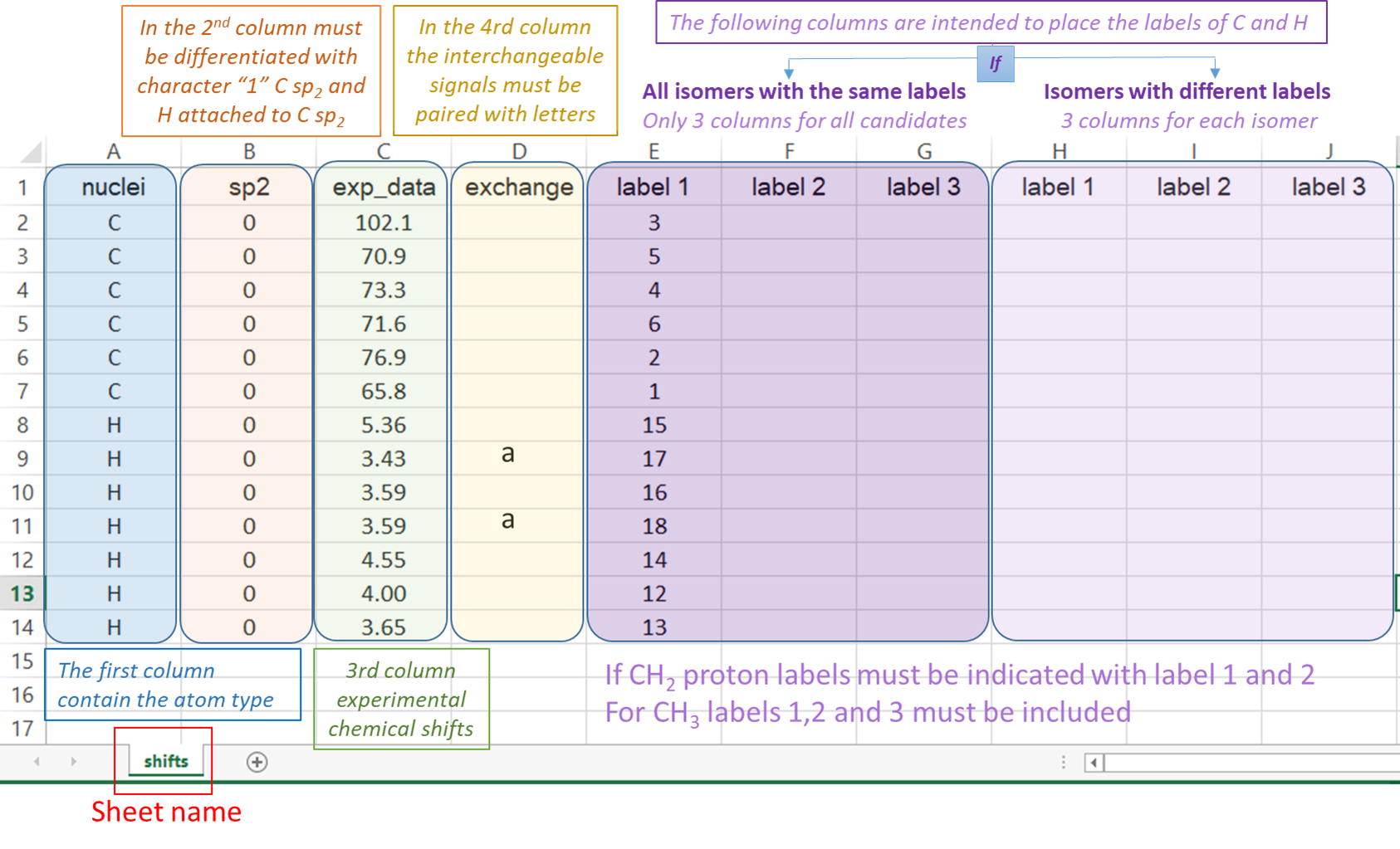
2) The input Excel file: The experimental data and the labels of the candidate structures must be placed in an excel file following the next rules. The excel file should be constituted by one sheet; containing the data of the NMR chemical shifts (named ‘shifts’).
“shifts” sheet Structure: the first column “nuclei” contain the identity of the atom ‘c or C’ for 13C and ‘h or H’ for hydrogen atoms. The second column “sp2” serves to indicate 0 (for sp3 C or H attached to) or 1 (for sp2 and sp). The third column “exp_data” contains the experimental chemical shifts. The column “exchange” serves to indicate by any character experimental data interchangeable (for instance two diatereotopics H must be indicated by an “a” in this column, this will cause for each candidate both the experimental and calculated values to be ordered from highest to lowest. The following columns are intended to place the labels of the nuclei associated to the corresponding chemical shift. If two or more values are added in that region, the isotropic shielding values will be averaged (as in the case of methyl groups or equivalent methylene groups). In the cases where isomers have different labels, there should be three columns for each isomer as indicated below.
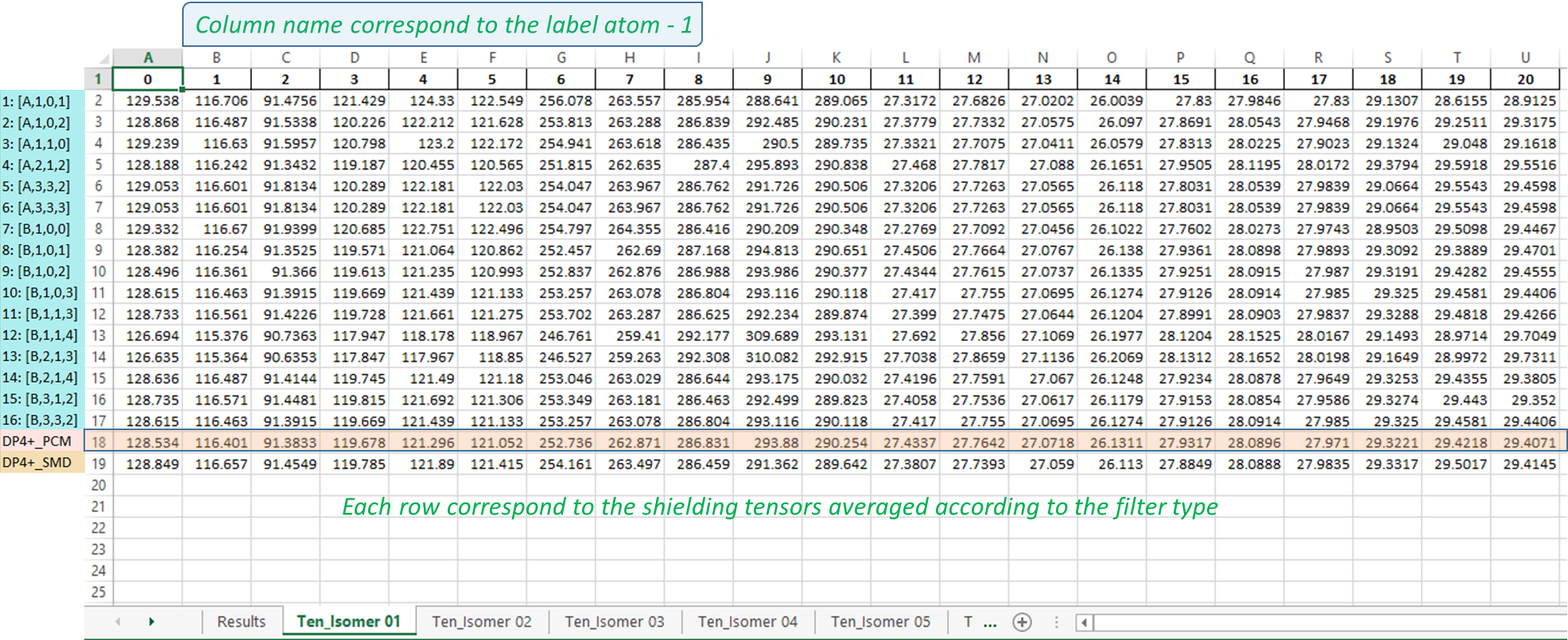
3) The output excel file: once the messi.py is executed, a file named ‘MESSI_Results.xlsx’ is created in the folder containing the Gaussian outputs. The file contains n+1 sheets where n is the number candidates structures:
Results sheet: this sheet contain the PCM-DP4+ (row 19, DP4+ standard), SMD-DP4+ (row 20, DP4+ standard but using SCF energies at the level SMD/B3LYP/6-31+G**), from row 3 to 18 you can find each of the 16 selected ensembles and in the second row it’s the average probability named MESSI. These probabilities are the full-DP4+ if both data 1H and 13C are available or it can be a partial probability, depending on whether data from H or C is used.
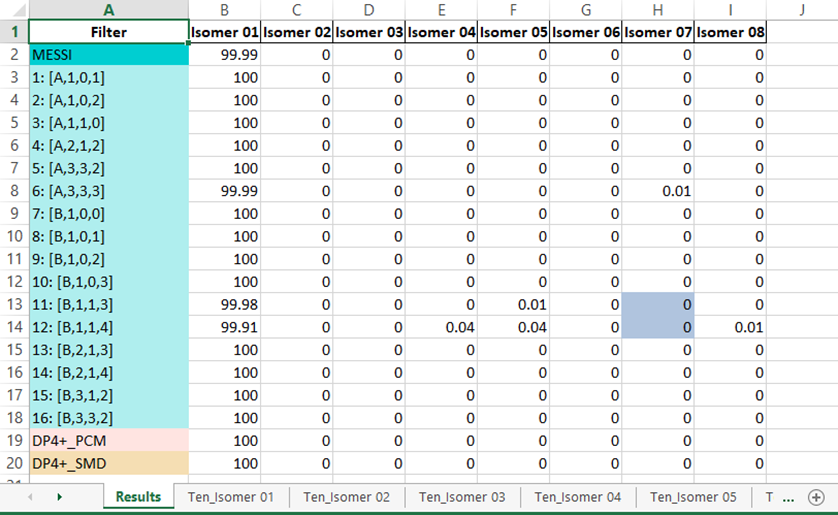
NOTE: It is important to point out that filters 4, 11 y 12 remove a fixed energy window of 1 Kcal from minimum, so if any isomer is left without conformations the probability will be 0 for that isomer. However, to notice of this situation, the cells corresponding to the isomer that did not participate in that ensemble will be indicated in gray.
Isomers tensors sheets: the excel file will contain as many sheets as candidate structures you modeled label as “Tens_Isomer N”, where N is the isomer number. Each sheet contains the weighted isotropic shielding constants according to the used ensemble; each assembly will be represented in a row in the same order as the probability results. As indicate in Figure.
Workflow and general recommendations
Step 1: Despite the new MESSI can handle any amount of isomers, keeping the number of candidates to a minimum has several advantages, as it reduces both the overall computational cost and the probability that the calculated data for an incorrect isomer ends up having better fit with the experimental values than the correct candidate.
Step 2: The conformational search should provide a good description of the conformational landscape of the system under study. Improper computational work might lead to potentially negative consequences in the overall results. Systematic sampling is always recommended, but impractical in highly flexible molecules. In those cases, stochastic searches using a reasonably large number of steps should be carried out. To avoid missing potentially relevant conformations, all conformations within a safe energy window from the corresponding global minimum should be kept. For this application, we recommend a 10 kcal/mol cutoff value using the MMFFaq force field.
Step 3: NMR and SCF calculation for all conformers of all candidate structures must be carried out at the levels PCM/mPW1PW91/6-31+G** and SMD/B3LYP/6-31+G** level respectively.
Step 4: The output files must be compiled in a folder. Additionally an Excel file with the experimental data and labels is needed.
Step 5: Run the script messi.py to perform the PCM-DP4+, SMD-DP4+ and MESSI probabilities calculations. The script will open a window where you can select the folder that contains the Gaussian output files, as *.log or *.out; and the Excel input file. The script feeds on the corresponding NMR and SCRF/SMD single point Gaussian output files. Both types of calculations could be running separately or together through the "link" option. The script automatically extract the isotropic shielding tensors and energies from each output and classifies them per isomer. Finally, the chemical shifts are averaged according the filter type and correlated with the experimental data to use it in the DP4+ formalism. The results are printed in an Excel file named ‘MESSI_Results.xlsx’.
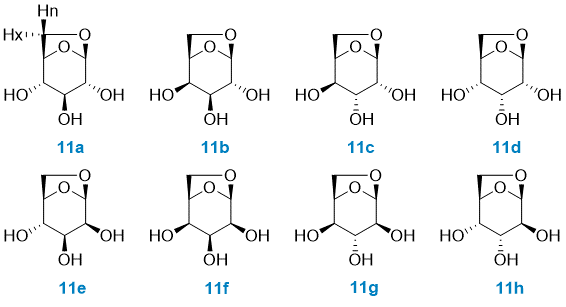
Case study: 1,6-anhydrohexopyranosides
In order to illustrate the MESSI workflow, we present the analysis of 1,6-anhydrohexopyranosides family. As indicated in the Figure, there are eight possible isomers.
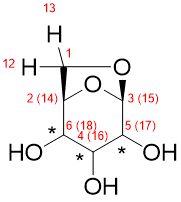
Following the recommended computational procedure, a total number of 130 conformers were found after the optimization at the B3LYP/6-31G* level (the standard for DP4+ calculations). Each structure was submitted to NMR and SCRF calculations at the PCM/mPW1PW91/6-31+G** and SMD/B3LYP/6-31+G** level respectively. The corresponding output files are provided in the Folder “Example”. According to Gaussian numbering scheme, the labels corresponding to each nuclei are given in Figure, Carbon label followed by its corresponding proton(s) label(s) between parenthesis.
MESSI Analysis
MESSI calculations were running for the eight possible distereoisomer, and placed in a folder. Once the script is run, the resulting excel report file “MESSI_Results” will be generated.
MESSI input and output excel files
Then execute messi_nmr and follow the steps.
Detail information is given in UserGuide.pdf included in the package. (Direct acces from GUI)
See the project repository for more details.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
Hashes for messi_nmr-0.1.12-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 12f8b76c7b4b2e691e62c0050e56cb100865a2cb2ad68930f2add31adfd92bf6 |
|
| MD5 | 1f3511496b71ab52398f2bd4911cc0e6 |
|
| BLAKE2b-256 | 770507c294adb5ad942ff50cae1711eeeaa863600fb9f8d9fe3559186f211d8c |