a plugin for mother machine image analysis
Project description
napari-mm3
A plugin for Mother Machine Image Analysis by Jun Lab.
This [napari] plugin was generated with [Cookiecutter] using [@napari]'s [cookiecutter-napari-plugin] template.
Installation
Ensure python and napari are installed in your system. To install the plugin, use:
pip install napari-mm3
Contributing
Contributions are very welcome. Tests can be run with [tox], please ensure the coverage at least stays the same before you submit a pull request.
Workflow
Generally, there is one widget for each process.
Basic workflow is as follows:
- Locate channels, create channel stacks, and return metadata (Compile widget).
- User guided selection of empty and full channels (ChannelSorter widget).
- Subtract phase contrast images (Subtract widget).
- Segment images (Segment widget).
- Create cell lineages
mm3 currently currently takes individual TIFF images as its input. If there are multiple color layers, then each TIFF image should be a stack of planes corresponding to a color. The quality of your images is important for mm3 to work properly.
The working directory now contains:
.
├── 20170720_SJ388_mopsgluc12aa.nd2
├── TIFF
1. Locate channels, create channel stacks, and return metadata (Compile widget).
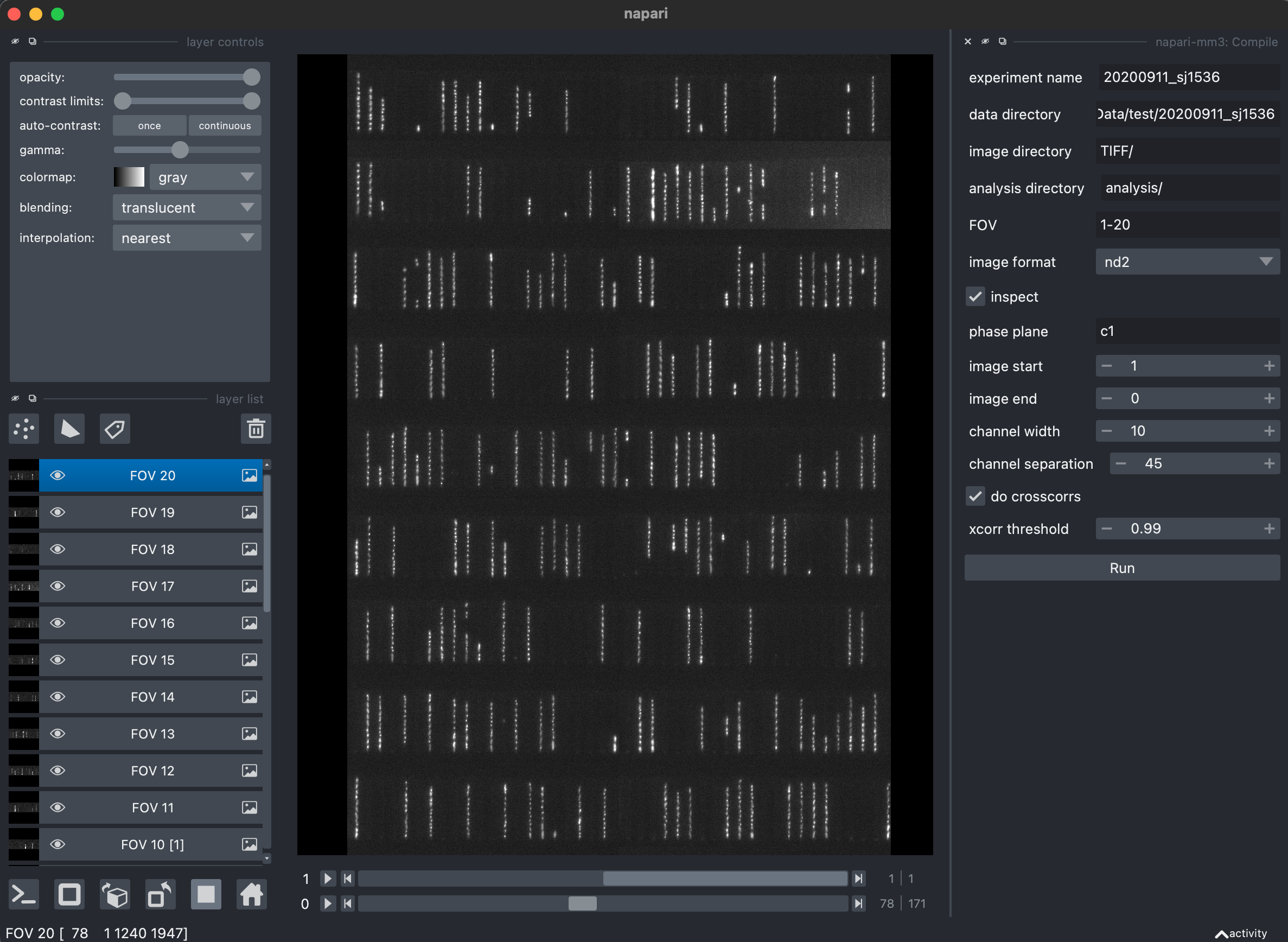
The Compile widget attempts to automatically identify and crop out individual growth channels. Images corresponding to a specific channel are then stacked in time, and these "channel stacks" are the basis of further analysis. If there are multiple colors, a channel stack is made for each color for each channel.
It is also at this time that metadata is drawn from the images and saved.
Parameters
TIFF_sourceneeds to be specified to indicate how the script should look for TIFF metadata. Choices areelementsandnd2ToTIFF.channel_width,channel_separation, andchannel_detection_snr, which are used to help find the channels.channel_length_padandchannel_width_padwill increase the size of your channel slices.t_end: Will only analyze images up to this time point. Useful for debugging.
The working directory now contains:
.
├── 20170720_SJ388_mopsgluc12aa.nd2
├── TIFF
├── analysis
│ ├── time_table.pkl
│ ├── time_table.txt
│ ├── TIFF_metadata.pkl
│ ├── TIFF_metadata.txt
│ ├── channel_masks.pkl
│ ├── channel_masks.txt
│ └── channels
2. User guided selection of empty and full channels (ChannelSorter).
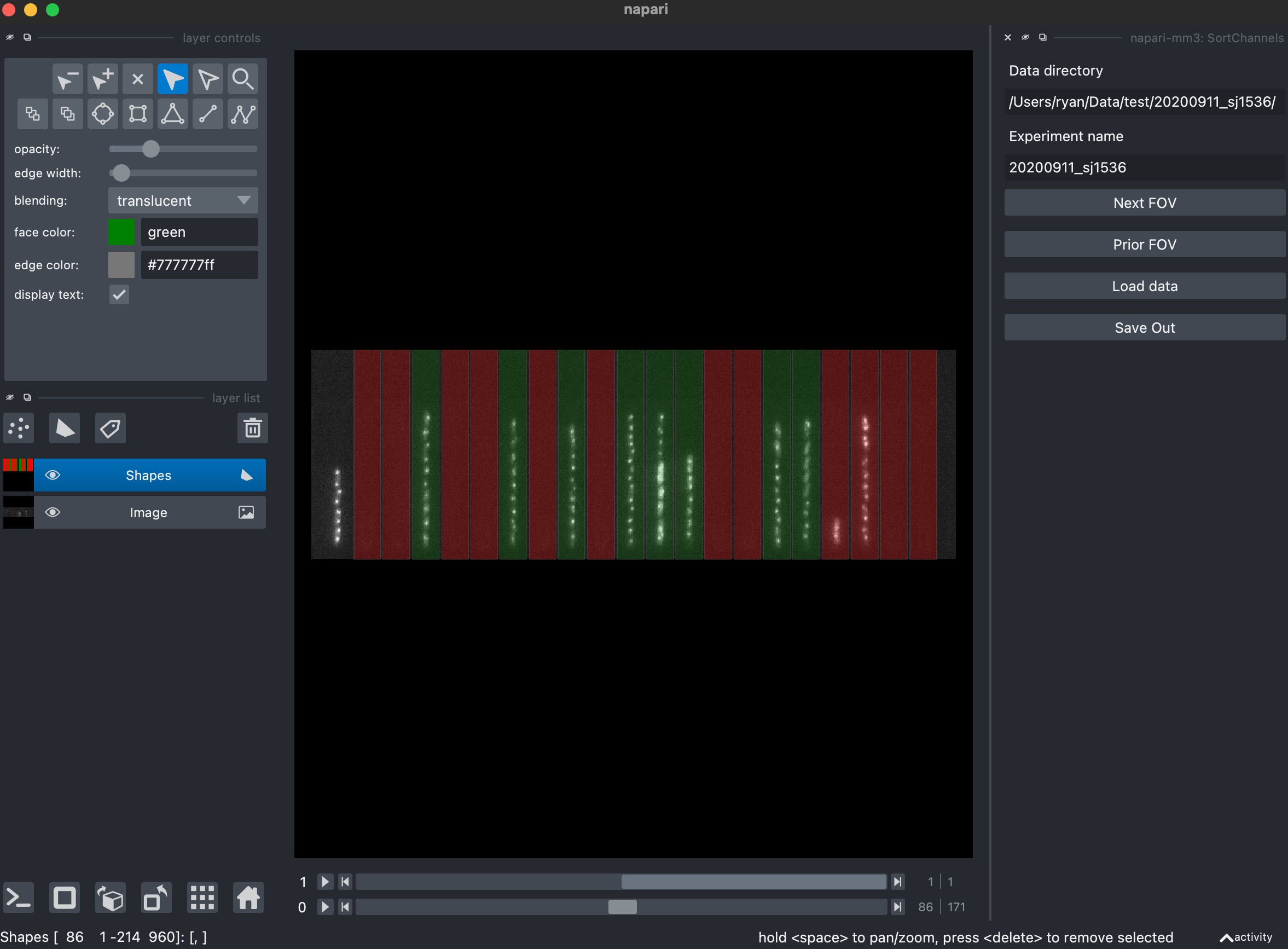
The Compile widget identifies all growth channels, regardless of if they contain or do not contain cells. ChannelSorter first attempts to guess, and then presents the user with a GUI to decide which channels should be analyzed, which channels should be ignored, and which channels should be used as empty channels during subtraction. This information is contained within the specs.yaml file.
Clicking the "Load data" button displays the first FOV analyzed, along with the program's predicted channel classification. The user is asked to click on the channels to change their designation between analyze (green), empty (blue) and ignore (red).
Click on the colored channels until they are as you wish. To navigate between fields of view click the "next FOV" or "prior FOV" buttons. The widget will output the specs file with channels indicated as analyzed (green, 1), empty for subtraction (blue, 0), or ignore (red, -1).
Parameters
phase_planeis the postfix of the channel which contains the phase imageschannel_picking_thresholdis a measure of correlation between a series of images, so a value of 1 would mean the same image over and over. Channels with values above this value (like empty channels) will be designated as empty before the user selection GUI.
The working directory is now:
.
├── 20170720_SJ388_mopsgluc12aa.nd2
├── TIFF
├── analysis
│ ├── time_table.pkl
│ ├── time_table.txt
│ ├── TIFF_metadata.pkl
│ ├── TIFF_metadata.txt
│ ├── channel_masks.pkl
│ ├── channel_masks.txt
│ ├── channels
│ ├── crosscorrs.pkl
│ ├── crosscorrs.txt
│ ├── specs.yaml
3. Subtract phase contrast images (Subtract widget).
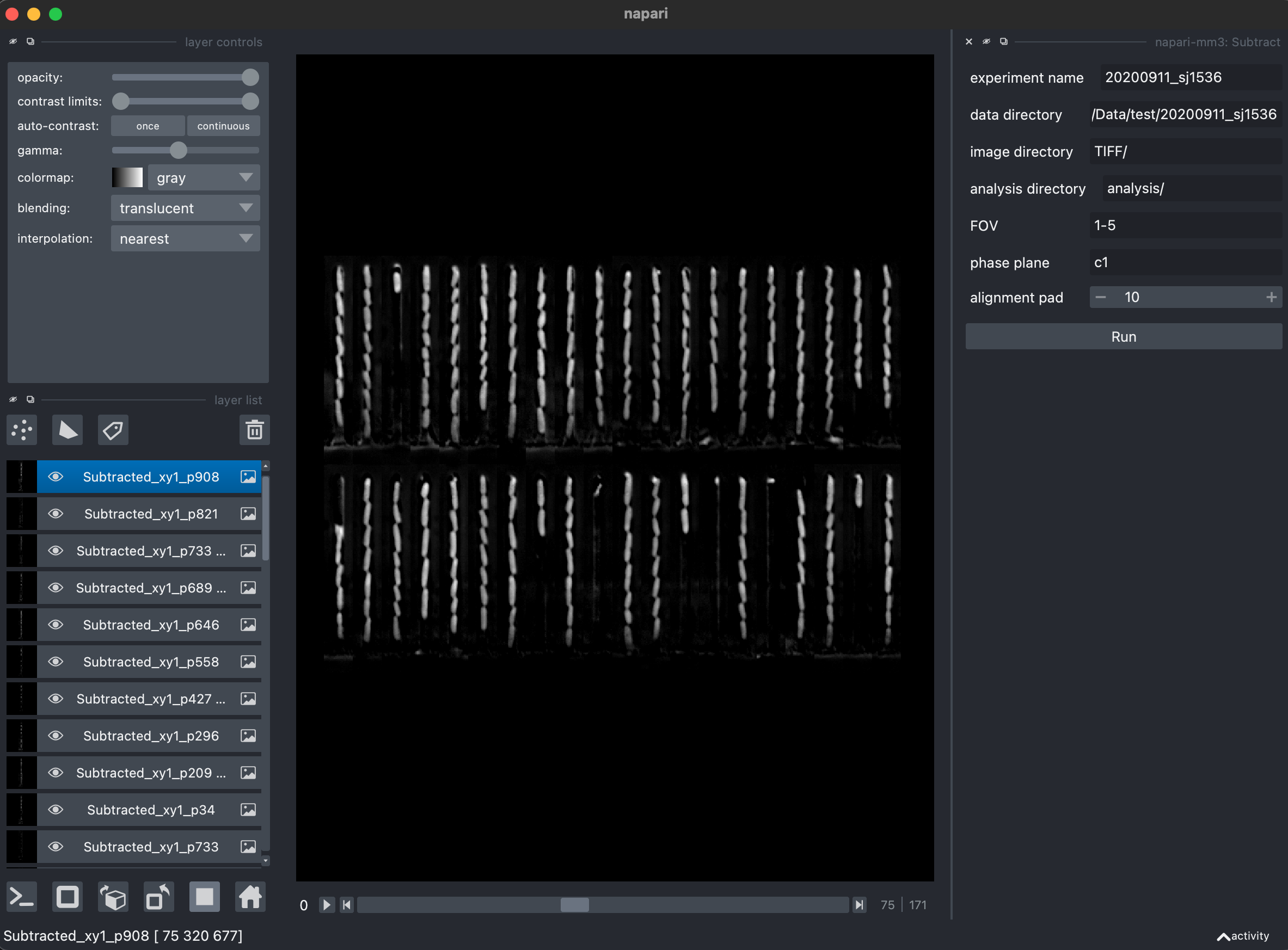
Downstream analysis of phase contrast (brightfield) images requires background subtraction to remove artifacts of the PDMS device in the images.
The working directory is now:
.
├── 20170720_SJ388_mopsgluc12aa.nd2
├── TIFF
├── analysis
│ ├── time_table.pkl
│ ├── time_table.txt
│ ├── TIFF_metadata.pkl
│ ├── TIFF_metadata.txt
│ ├── channel_masks.pkl
│ ├── channel_masks.txt
│ ├── channels
│ ├── crosscorrs.pkl
│ ├── crosscorrs.txt
│ ├── empties
│ ├── specs.yaml
│ └── subtracted
4. Segment images (SegmentOTSU or SegmentUnet).
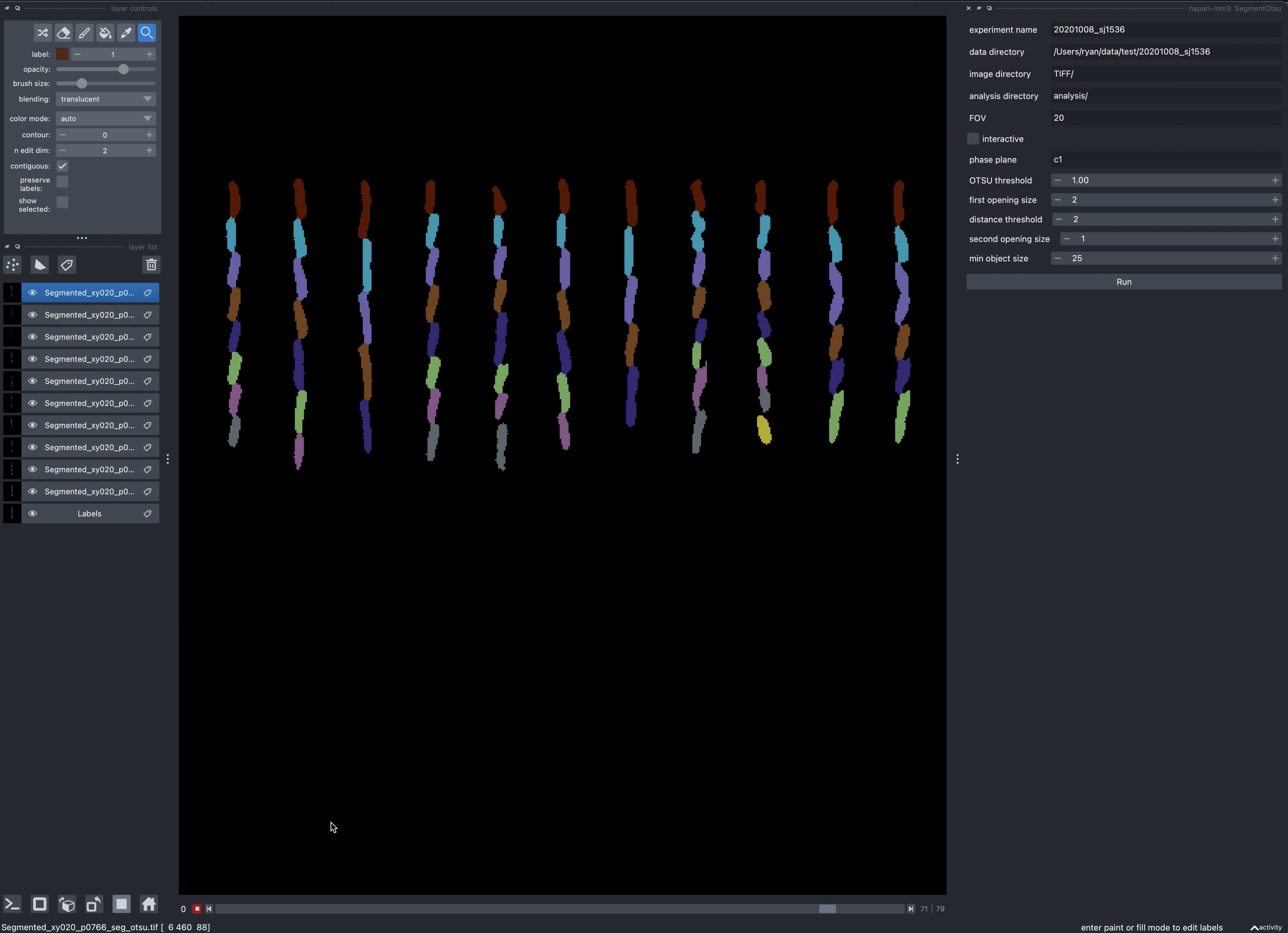
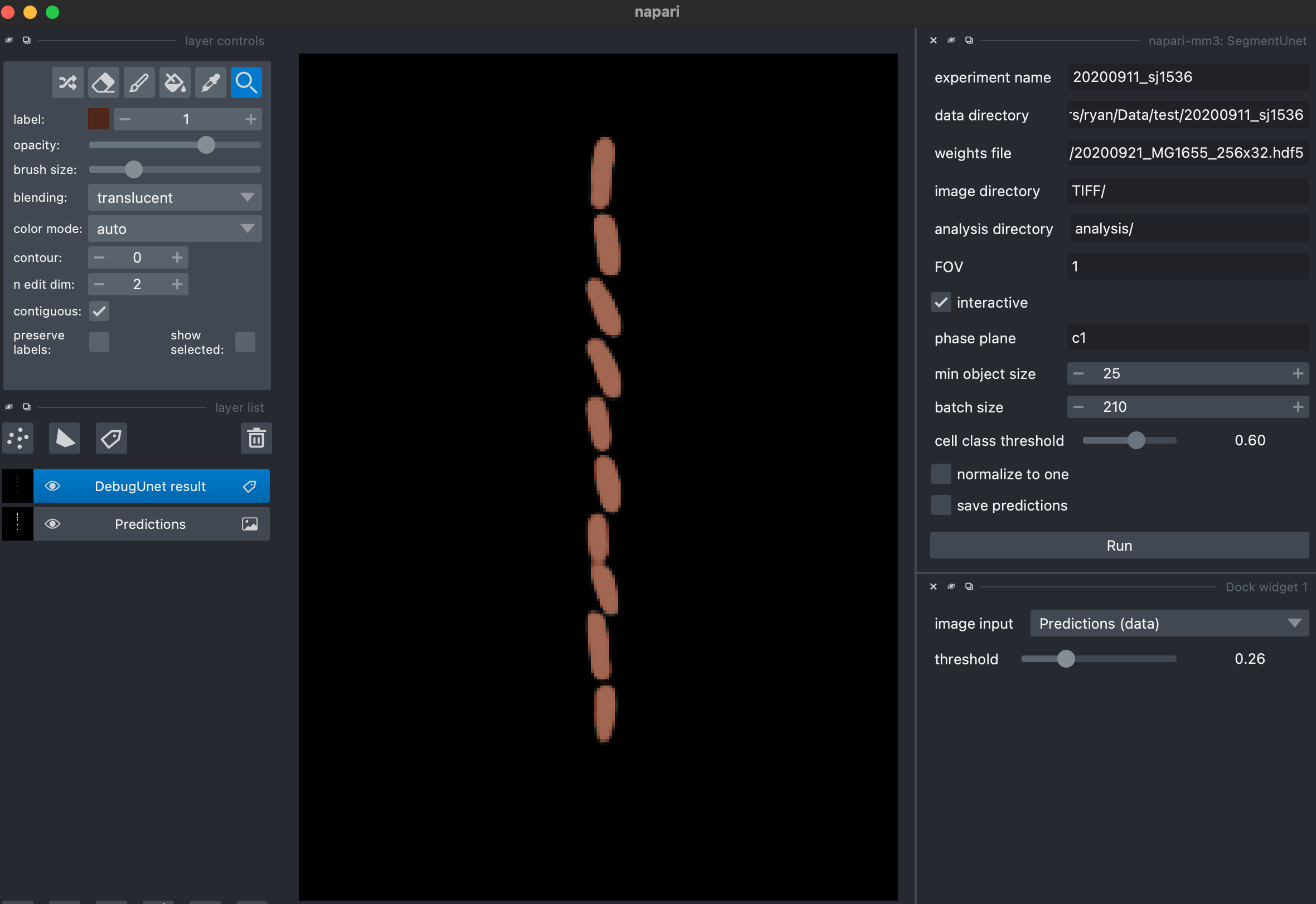 mm3 can use either deep learning or a traditional machine vision approach (Otsu thresholding, morphological operations and watershedding) to locate cells from the subtracted images.
mm3 can use either deep learning or a traditional machine vision approach (Otsu thresholding, morphological operations and watershedding) to locate cells from the subtracted images.
OTSU parameters
first_opening_size: Size in pixels of first morphological opening during segmentation.distance_threshold: Distance in pixels which thresholds distance transform of binary cell image.second_opening_size: Size in pixels of second morphological opening.
U-net parameters
threshold: threshold value (between 0 and 1) for cell classificationmin_object_size: Objects smaller than this area in pixels will be removed before labeling.
The working directory is now:
.
├── 20170720_SJ388_mopsgluc12aa.nd2
├── TIFF
├── analysis
│ ├── time_table.pkl
│ ├── time_table.txt
│ ├── TIFF_metadata.pkl
│ ├── TIFF_metadata.txt
│ ├── channel_masks.pkl
│ ├── channel_masks.txt
│ ├── channels
│ ├── crosscorrs.pkl
│ ├── crosscorrs.txt
│ ├── empties
│ ├── segmented
│ ├── specs.yaml
│ └── subtracted
5. Create cell lineages (Track widget).
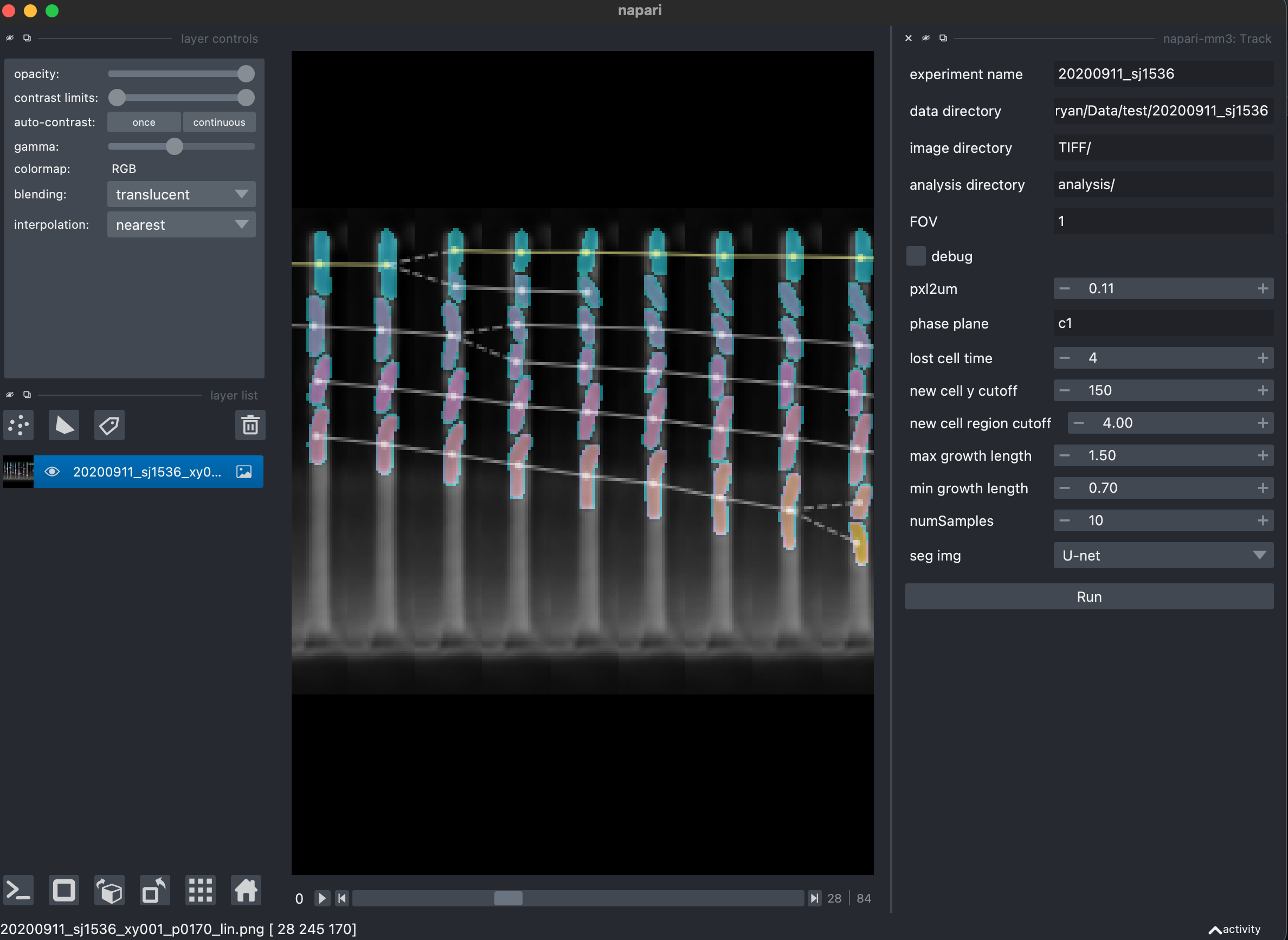
After cells are found for each channel in each time point, these labeled cells are connected across time to create complete cells and lineages.
Parameters
lost_cell_time: If this many time points pass and a region has not yet been linked to a future region, it is dropped.max_growth_length: If a region is to be connected to a previous region, it cannot be larger in length by more than this ratio.min_growth_length: If a region is to be connected to a previous region, it cannot be smaller in length by less than this ratio.max_growth_area: If a region is to be connected to a previous region, it cannot be larger in area by more than this ratio.min_growth_area: If a region is to be connected to a previous region, it cannot be smaller in area by less than this ratio.new_cell_y_cutoff: distance in pixels from closed end of image above which new regions are not considered for starting new cells.segmentation_method: whether to construct lineage from cells segmented by the Otsu or U-net method
The working directory is now:
.
├── 20170720_SJ388_mopsgluc12aa.nd2
├── TIFF
├── analysis
│ ├── time_table.pkl
│ ├── time_table.txt
│ ├── TIFF_metadata.pkl
│ ├── TIFF_metadata.txt
│ ├── cell_data
│ │ └── complete_cells.pkl
│ ├── channel_masks.pkl
│ ├── channel_masks.txt
│ ├── channels
│ ├── crosscorrs.pkl
│ ├── crosscorrs.txt
│ ├── empties
│ ├── segmented
│ ├── specs.pkl
│ ├── specs.txt
│ └── subtracted
└── params.yaml
## License
Distributed under the terms of the [BSD-3] license,
"napari-mm3" is free and open source software
## Issues
If you encounter any problems, please [file an issue] along with a detailed description.
[napari]: https://github.com/napari/napari
[Cookiecutter]: https://github.com/audreyr/cookiecutter
[@napari]: https://github.com/napari
[MIT]: http://opensource.org/licenses/MIT
[BSD-3]: http://opensource.org/licenses/BSD-3-Clause
[GNU GPL v3.0]: http://www.gnu.org/licenses/gpl-3.0.txt
[GNU LGPL v3.0]: http://www.gnu.org/licenses/lgpl-3.0.txt
[Apache Software License 2.0]: http://www.apache.org/licenses/LICENSE-2.0
[Mozilla Public License 2.0]: https://www.mozilla.org/media/MPL/2.0/index.txt
[cookiecutter-napari-plugin]: https://github.com/napari/cookiecutter-napari-plugin
[file an issue]: https://github.com/ahirsharan/napari-mm3/issues
[napari]: https://github.com/napari/napari
[tox]: https://tox.readthedocs.io/en/latest/
[pip]: https://pypi.org/project/pip/
[PyPI]: https://pypi.org/
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for napari_mm3-0.0.6-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 190a32e2fc086193a7556c9142d40a69675dd845f909bf2f0cbe3dc032acc166 |
|
| MD5 | 6451873ac6014eebb74f9db8cd7c34f7 |
|
| BLAKE2b-256 | 47cdf0b4c44f5d0832cf21b07cdc1ab3900c57e7f04449f72d6f51b3f9a12572 |


















