Omnipy is a high level Python library for type-driven data wrangling and scalable workflow orchestration (under development)
Project description
Omnipy is the new name of the Python package formerly known as uniFAIR.
We are very grateful to Dr. Jamin Chen, who gracefully transferred ownership of the (mostly unused) "omnipy" name in PyPI to us!
--
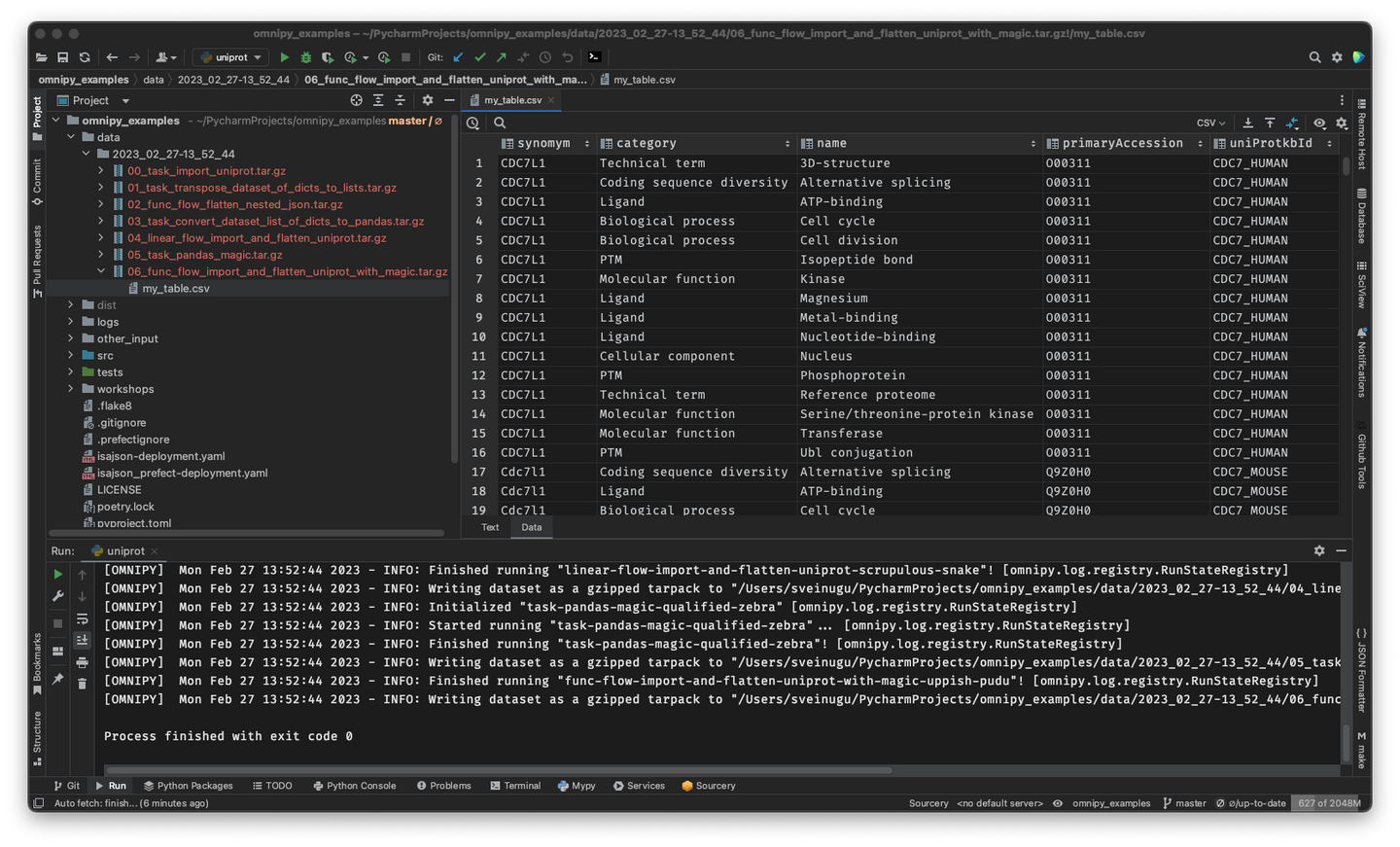
Update Feb 3, 2023: Documentation of the Omnipy API is still sparse. However, for running examples, please check out the omnipy-examples repo and its related PYPI package!
(NOTE: Read the section Transformation on the FAIRtracks.net website for a more detailed and better formatted version of the following description!)
Generic functionality
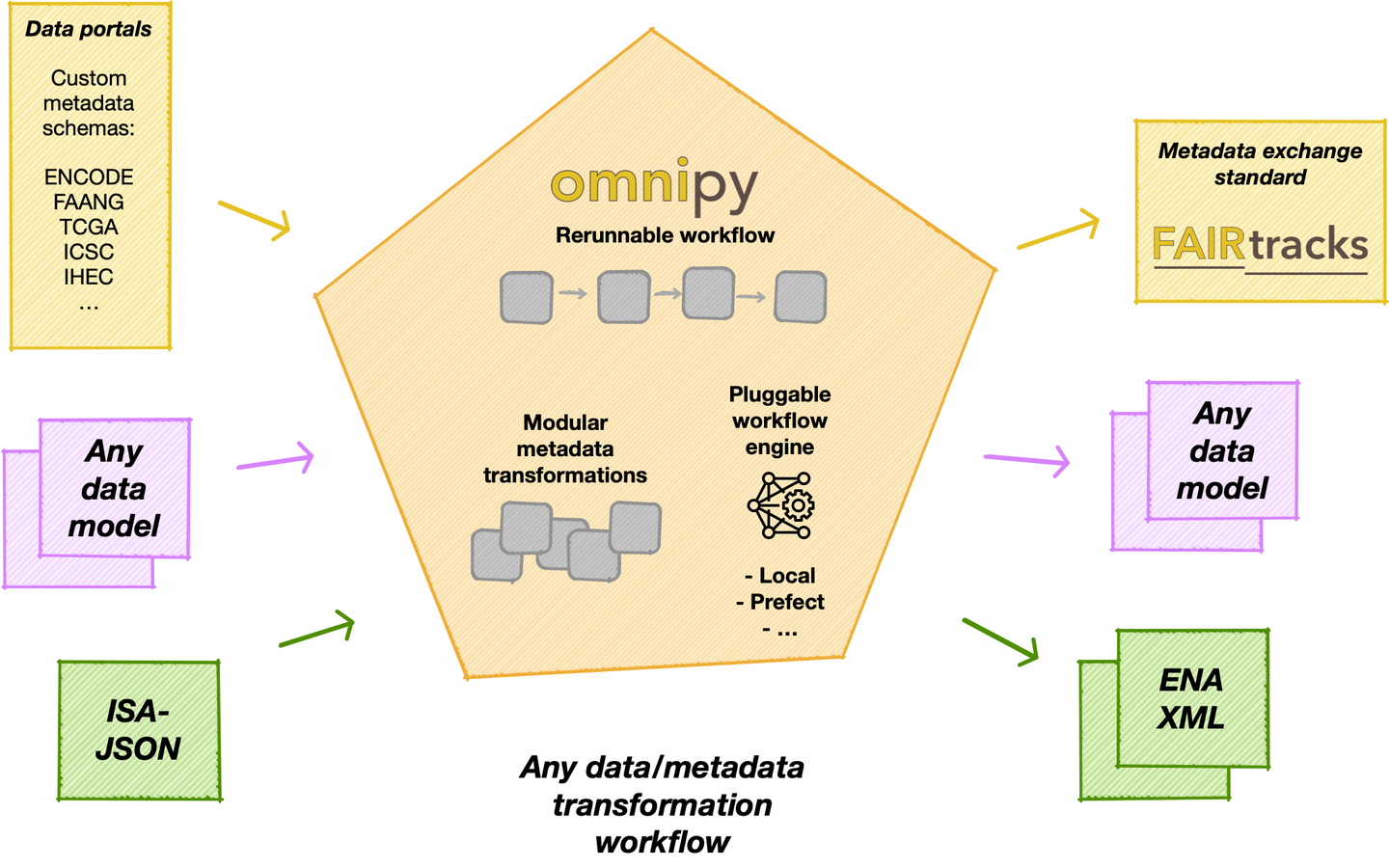
Omnipy is designed primarily to simplify development and deployment of (meta)data transformation processes in the context of FAIRification and data brokering efforts. However, the functionality is very generic and can also be used to support research data (and metadata) transformations in a range of fields and contexts beyond life science, including day-to-day research scenarios:
Data wrangling in day-to-day research: Researchers in life science and other data-centric fields often need to extract, manipulate and integrate data and/or metadata from different sources, such as repositories, databases or flat files. Much research time is spent on trivial and not-so-trivial details of such "data wrangling":
- reformat data structures
- clean up errors
- remove duplicate data
- map and integrate dataset fields
- etc.
General software for data wrangling and analysis, such as Pandas, R or Frictionless, are useful, but researchers still regularly end up with hard-to-reuse scripts, often with manual steps.
Step-wise data model transformations: With the Omnipy Python package, researchers can import (meta)data in almost any shape or form: nested JSON; tabular (relational) data; binary streams; or other data structures. Through a step-by-step process, data is continuously parsed and reshaped according to a series of data model transformations.
"Parse, don't validate": Omnipy follows the principles of "Type-driven design" (read Technical note #2: "Parse, don't validate" on the FAIRtracks.net website for more info). It makes use of cutting-edge Python type hints and the popular pydantic package to "pour" data into precisely defined data models that can range from very general (e.g. "any kind of JSON data", "any kind of tabular data", etc.) to very specific (e.g. "follow the FAIRtracks JSON Schema for track files with the extra restriction of only allowing BigBED files").
Data types as contracts: Omnipy tasks (single steps) or flows (workflows) are defined as transformations from specific input data models to specific output data models. pydantic-based parsing guarantees that the input and output data always follows the data models (i.e. data types). Thus, the data models defines "contracts" that simplifies reuse of tasks and flows in a mix-and-match fashion.
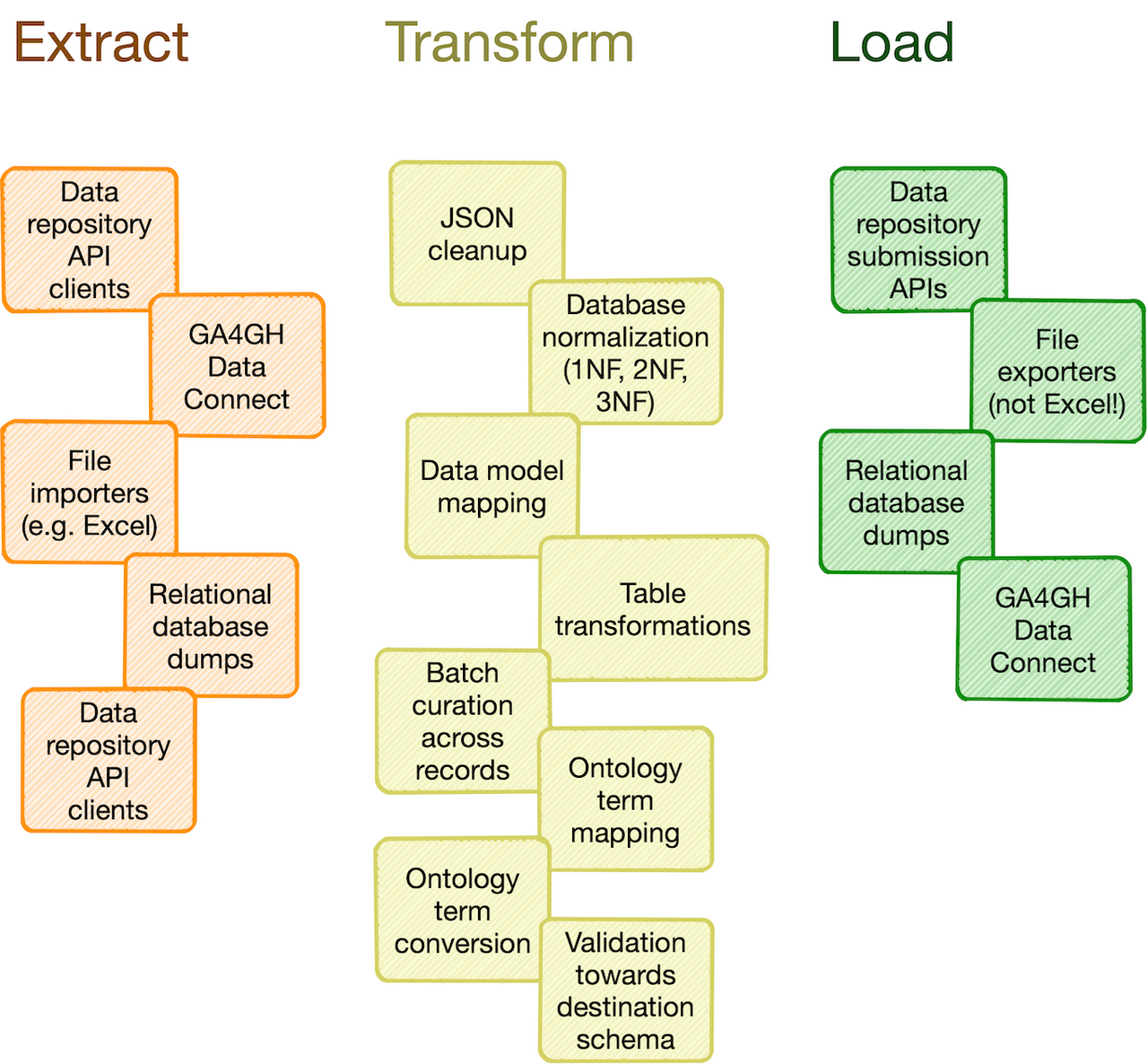
Catalog of common processing steps: Omnipy is built from the ground up to be modular. We aim to provide a catalog of commonly useful functionality ranging from:
- data import from REST API endpoints, common flat file formats, database dumps, etc.
- flattening of complex, nested JSON structures
- standardization of relational tabular data (i.e. removing redundancy)
- mapping of tabular data between schemas
- lookup and mapping of ontology terms
- semi-automatic data cleaning (through e.g. Open Refine)
- support for common data manipulation software and libraries, such as Pandas, R, Frictionless, etc.
In particular, we will provide a FAIRtracks module that contains data models and processing steps to transform metadata to follow the FAIRtracks standard.
Refine and apply templates: An Omnipy module typically consists of a set of generic task and flow templates with related data models, (de)serializers, and utility functions. The user can then pick task and flow templates from this extensible, modular catalog, further refine them in the context of a custom, use case-specific flow, and apply them to the desired compute engine to carry out the transformations needed to wrangle data into the required shape.
Rerun only when needed: When piecing together a custom flow in Omnipy, the user has persistent access to the state of the data at every step of the process. Persistent intermediate data allows for caching of tasks based on the input data and parameters. Hence, if the input data and parameters of a task does not change between runs, the task is not rerun. This is particularly useful for importing from REST API endpoints, as a flow can be continuously rerun without taxing the remote server; data import will only carried out in the initial iteration or when the REST API signals that the data has changed.
Scale up with external compute resources: In the case of large datasets, the researcher can set up a flow based on a representative sample of the full dataset, in a size that is suited for running locally on, say, a laptop. Once the flow has produced the correct output on the sample data, the operation can be seamlessly scaled up to the full dataset and sent off in software containers to run on external compute resources, using e.g. Kubernetes. Such offloaded flows can be easily monitored using a web GUI.
Industry-standard ETL backbone: Offloading of flows to external compute resources is provided by the integration of Omnipy with a workflow engine based on the Prefect Python package. Prefect is an industry-leading platform for dataflow automation and orchestration that brings a series of powerful features to Omnipy:
- Predefined integrations with a range of compute infrastructure solutions
- Predefined integration with various services to support extraction, transformation, and loading (ETL) of data and metadata
- Code as workflow ("If Python can write it, Prefect can run it")
- Dynamic workflows: no predefined Direct Acyclic Graphs (DAGs) needed!
- Command line and web GUI-based visibility and control of jobs
- Trigger jobs from external events such as GitHub commits, file uploads, etc.
- Define continuously running workflows that still respond to external events
- Run tasks concurrently through support for asynchronous tasks
Pluggable workflow engines: It is also possible to integrate Omnipy with other workflow backends by implementing new workflow engine plugins. This is relatively easy to do, as the core architecture of Omnipy allows the user to easily switch the workflow engine at runtime. Omnipy supports both traditional DAG-based and the more avant garde code-based definition of flows. Two workflow engines are currently supported: local and prefect.
Scenarios
As initial use cases, we will consider the following two scenarios:
- Transforming ENCODE metadata into FAIRtracks format
- Transforming TCGA metadata into FAIRtracks format
Nomenclature:
- Omnipy is designed to work with content which could be classified both as data and metadata in their original context. For simplicity, we will refer to all such content as "data".
Overview of the proposed FAIRification process:
-
Step 1: Import data from original source:
-
1A: From API endpoints
- Input: API endpoint producing JSON data
- Output: JSON files (possibly with embedded JSON objects/lists [as strings])
- Description: General interface to support various API endpoints. Import all data by crawling API endpoints providing JSON content
- Generalizable: Partly (at least reuse of utility functions)
- Manual/automatic: Automatic
- Details:
- GDC/TCGA substeps (implemented as Step objects with file input/output)
-
1A. Filtering step:
- Input: parameters defining how to filter, e.g.:
- For all endpoints (projects, cases, files, annotations), support:
- Filter on list of IDs
- Specific number of items
- All
- Example config:
- projects: 2 items
- cases: 2 items
- files: all
- annotations: all
- Define standard configurations, e.g.:
- Default: limited extraction (3 projects * 3 cases * 5 files? (+ annotations?))
- All TCGA
- List of projects
- Hierarchical for loop through endpoints to create filter definitions
- For all endpoints (projects, cases, files, annotations), support:
- Output: Filter definitions as four files, e.g. as JSON,
as they should be input to the filter parameter to the API:
projects_filter.json: { "op": "in", "content": { "field": "project_id", "value": ['TCGA_ABCD', 'TCGA_BCDE'] } } cases_filter.json: { "op": "in", "content": { "field": "case_id", "value": ['1234556', '234567', '3456789', '4567890'] } } files_filter.json: { "op": "in", "content": { "field": "file_id", "value": ['1234556', '234567', '3456789', '4567890'] } } annotations.json { "op": "in", "content": { "field": "annotation_id", "value": ['1234556', '234567', '3456789', '4567890'] } }
- Input: parameters defining how to filter, e.g.:
-
1B. Fetch and divide all fields step:
-
Input: None
-
Output: JSON files specifying all the fields of an endpoint fetched from the
mappingAPI. The fields should be divided into chunks of a size that is small enough for the endpoints to handle. The JSON output should also specify the primary_key field, that needs to be added to all the API calls in order for the results to be joinable.Example JSON files:
projects_fields.json: { "primary_key": "project_id", "fields_divided": [ ["field_a", "field_b"], ["field_c.subfield_a", "field_c.subfield_b", "field_d"] ] } (...) # For all endpoints
-
-
1C. Download from all endpoints according to the filters and the field divisions. If there is a limitation on the number of hits that the endpoint is able to return, divide into smaller API calls for a certain number of hits and concatenate the results. Make sure that proper waiting time (1 second?) is added between the calls (to not overload the endpoint).
-
1D. Extract identifiers from nested objects (when present) and insert into parents objects
-
- ENCODE:
- Identify where to start (Cart? Experiment?)
- To get all data for a table (double-check this):
https://www.encodeproject.org/experiments/@@listing?format=json&frame=object - Download all tables directly.
- GDC/TCGA substeps (implemented as Step objects with file input/output)
-
1b: From JSON files
- Input: JSON content as files
- Output: Pandas DataFrames (possibly with embedded JSON objects/lists)
- Description: Import data from files. Requires specific parsers to be implemented.
- Generalizable: Fully
- Manual/automatic: Automatic
-
1c: From non-JSON files
- Input: File content in some supported format (e.g. GSuite)
- Output: Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- Description: Import data from files. Requires specific parsers to be implemented.
- Generalizable: Partly (generating reference metadata might be tricky)
- Manual/automatic: Automatic
-
1d: From database
- Input: Direct access to relational database
- Output: Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- Description: Import data from database
- Generalizable: Fully
- Manual/automatic: Automatic
-
-
Step 2: JSON cleanup
- Input: Pandas DataFrames (possibly with embedded JSON objects/lists)
- Output: Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- Description: Replace embedded objects with identifiers (possibly as lists)
- Generalizable: Partly (generating reference metadata might be tricky)
- Manual/automatic: Depending on original input
- Details:
- If there are embedded objects from other tables:
- ENCODE update:
- By using the
frame=objectparameter, we will not get any embedded objects from the APIs for the main tables. There is, however, some "auditing" fields that contain JSON objects. We can ignore these in the first iteration.
- By using the
- If the original table of the embedded objects can be retrieved directly from an API, replace such embedded objects with unique identifiers to the object in another table (maintaining a reference to the name of the table, if needed)
- Record the reference metadata
(table_from, attr_from) -> (table_to, attr_to)for joins:- Example:
(table: "experiment", column: "replicates") -> (table: "replicate", column: "@id")
- Example:
- Record the reference metadata
- If the original table of the embedded objects are not directly available from an API, one needs to fill out the other table with the content that is embedded in the current object, creating the table if needed.
- ENCODE update:
- For all fields with identifiers that reference other tables:
- Record the reference metadata
(table_from, attr_from) -> (table_to, attr_to)for joins. - If the field contains a list of identifiers
- Convert into Pandas Series
- Record the reference metadata
- If there are embedded objects from other tables:
-
Step 3: Create reference tables to satisfy 1NF
- Input: Pandas DataFrames (possibly containing lists of identifiers as Pandas Series) + reference metadata
- Output: Pandas DataFrames (original tables without reference column) [1NF] + reference tables + reference metadata
- Description: Move references into separate tables, transforming the tables in first normal form (1NF)
- Generalizable: Fully
- Manual/automatic: Automatic
- Details:
- For each reference pair:
- Create a reference table
- For each item in the "from"-reference column:
- Add new rows in the reference table for each "to"-identifier, using the same "from"-identifier
- Example: Table "experiment-replicate" with columns "experiment.@id", "replicate.@id"
- Add new rows in the reference table for each "to"-identifier, using the same "from"-identifier
- Delete the complete column from the original table
- For each reference pair:
-
Step 4: Satisfy 2NF
- Input: Pandas DataFrames (original tables without reference column) [1NF] + reference tables
- Output: Pandas DataFrames (original tables without reference column) [2NF] + reference tables
- Description: Automatic transformation of original tables into second normal form (2NF):
- Generalizable: Fully (if not, we skip it)
- Manual/automatic: Automatic
- Details:
- Use existing library.
-
Step 5: Satisfy 3NF
- Input: Pandas DataFrames (original tables without reference column) [2NF] + reference tables
- Output: Pandas DataFrames (original tables without reference column) [3NF] + reference tables
- Description: Automatic transformation of original tables into third normal form (3NF):
- Generalizable: Fully (if not, we skip it)
- Manual/automatic: Automatic
- Details:
- Use existing library.
-
Step 6: Create model map
- Input: Pandas DataFrames (original tables without reference column) [Any NF] + reference tables + FAIRtracks JSON schemas
- Output: Model map [some data structure (to be defined) mapping FAIRtracks objects and attributes to tables/columns in the original data]
- Description: Manual mapping of FAIRtracks objects and attributes to corresponding tables and columns in the original data.
- Generalizable: Fully
- Manual/automatic: Manual
- Details:
- For each FAIRtracks object:
- Define a start table in the original data
- For each FAIRtracks attribute:
- Manually find the path (or paths) to the original table/column that this maps to
- Example:
Experiments:organism (FAIRtracks) -> Experiments.Biosamples.Organism.scientific_name
- Example:
- Manually find the path (or paths) to the original table/column that this maps to
- For each FAIRtracks object:
-
Step 7: Apply model map to generate initial FAIRtracks tables
- Input: Pandas DataFrames (original tables without reference column) [Any NF] + reference tables + Model map
- Output: Pandas DataFrames (initial FAIRtracks tables, possibly with multimapped attributes)
- Example:
Experiment.target_from_origcolumn1andExperimentl.target_from_origcolumn2contain content from two different attributes from the original data that both corresponds toExperiment.target
- Example:
- Description: Generate initial FAIRtracks tables by applying the model map, mapping FAIRtracks attributes with one or more attributes (columns) in the original table.
- Generalizable: Fully
- Manual/automatic: Automatic
- Details:
- For every FAIRtracks object:
- Create a new pandas DataFrame
- For every FAIRtracks attribute:
- From the model map, get the path to the corresponding original table/column, or a list of such paths in case of multimapping
- For each path:
- Automatically join tables to get primary keys and attribute value in the same table:
- Example:
experiment-biosample JOIN biosample-organism JOIN organismwill create mapping table with two columns:Experiments.local_idandOrganism.scientific_name
- Example:
- Add column to FAIRtracks DataFrame
- In case of multimodeling, record the relation between FAIRtracks attribute and corresponding multimapped attributes, e.g. by generating unique attribute names for each path, such as
Experiment.target_from_origcolumn1andExperiment.target_from_origcolumn2, which one can derive directly from the model map.
- Automatically join tables to get primary keys and attribute value in the same table:
- For every FAIRtracks object:
-
Step 8: Harmonize multimapped attributes
- Input: Pandas DataFrames (initial FAIRtracks tables, possibly with multimapped attributes) + model map
- Output: Pandas DataFrames (initial FAIRtracks tables)
- Description: Harmonize multimapped attributes manually, or possibly by applying scripts
- Generalizable: Limited (mostly by reusing util functions)
- Manual/automatic: Mixed (possibly scriptable)
- Details:
- For all multimapped attributes:
- Manually review values (in batch mode) and generate a single output value for each combination:
- Hopefully Open Refine can be used for this. If so, one needs to implement data input/output mechanisms.
- Manually review values (in batch mode) and generate a single output value for each combination:
- For all multimapped attributes:
-
Further steps to be detailed:
- For all FAIRtracks attributes with ontology terms: Convert terms using required ontologies
- Other FAIRtracks specific value conversion
- Manual batch correction of values (possibly with errors), probably using Open Refine
- Validation of FAIRtracks document
Suggestion: we will use Pandas DataFrames as the core data structure for tables, given that the library provides the required features (specifically Foreign key and Join capabilities)
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.