Unsupervised learning of the structure of particulate systems
Project description
partycls
partycls is a Python package for spatio-temporal cluster analysis of interacting particles. It provides descriptors suitable for applications in condensed matter physics and integrates the necessary tools of unsupervised learning into a streamlined workflow. Thanks to a flexible system of filters, it makes it easy to restrict the analysis to a given subset of particles based on arbitrary particle properties.
Quick start
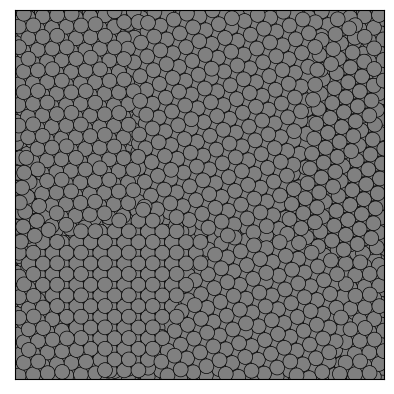
Here is a simple example that shows how to use partycls to find identify grain boundaries in a polycrystalline system. The system configuration is stored in a trajectory file with a single frame
from partycls import Trajectory
traj = Trajectory('grains.xyz')
traj[0].show()
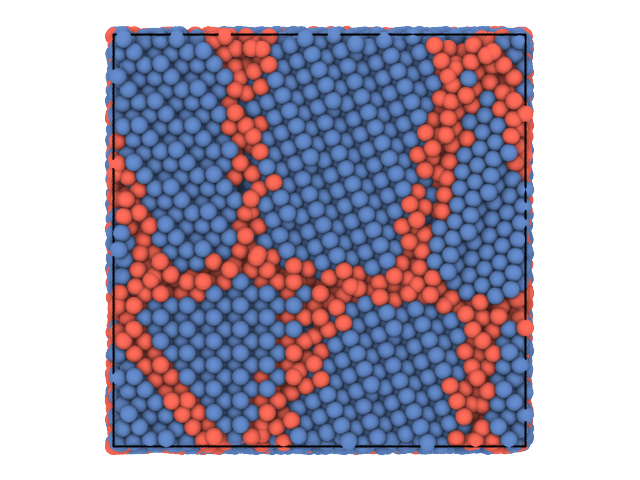
We use the local distribution of bond angles around each particle as a structural fingerprint and perform a clustering using the K-Means algorithm. We show the system again, this time coloring the particles according to the cluster they belong to.
from partycls import Workflow
wf = Workflow(traj, descriptor='ba', clustering='kmeans')
wf.run()
traj[0].show(color='label')
Tha results are also written to a set of files including a labeled trajectory file and additional information on the clustering results. The whole workflow can be easily tuned and customized, check out the tutorials to see how and for further examples.
We can restrict the analysis to specific a subset of particles by adding filters. Say we have a binary mixture composed of particles with types A and B, and are only interested in the angular correlations of B particles in the left side of the box (with respect to x-axis):
from partycls import Trajectory
from partycls.descriptor import BondAngleDescriptor
traj = Trajectory('trajectory.xyz')
D = BondAngleDescriptor(traj)
D.add_filter("species == 'B'")
D.add_filter("x < 0.0")
D.compute()
# Angular correlations for the selected particles
print(D.features)
We can then perform a clustering based on these structural features, asking for e.g. 3 clusters:
from partycls import KMeans
clustering = KMeans(n_clusters=3)
clustering.fit(D.features)
print('Cluster membership of the particles', clustering.labels)
Features
partycls is designed to accept a large variety of trajectory formats (including custom ones!) either through its built-in trajectory reader or via third-party packages, such as MDTraj and atooms. It relies on the scikit-learn package to perform feature scaling as well as a number of dimensionality reduction and clustering methods. In addition to its native descriptors, partycls supports additional structural descriptors via DScribe.
Requirements
- numpy
- scikit-learn
- [optional] mdtraj (additional trajectory formats)
- [optional] atooms (additional trajectory formats)
- [optional] dscribe (additional descriptors)
- [optional] py3Dmol (interactive 3D visualization)
Documentation
See the tutorials (Jupyter notebooks) for a step-by-step introduction to the main features of partycls and some of its applications.
Installation
From pypi:
pip install partycls
From the code repository:
git clone https://github.com/jorisparet/partycls.git
cd partycls
make install
Authors
Joris Paret
Daniele Coslovich: http://www-dft.ts.infn.it/~coslovich/
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.