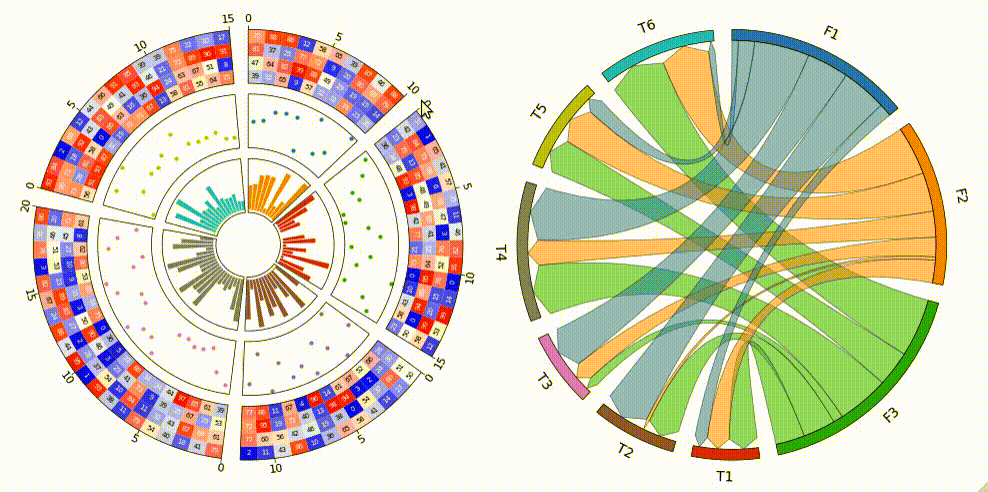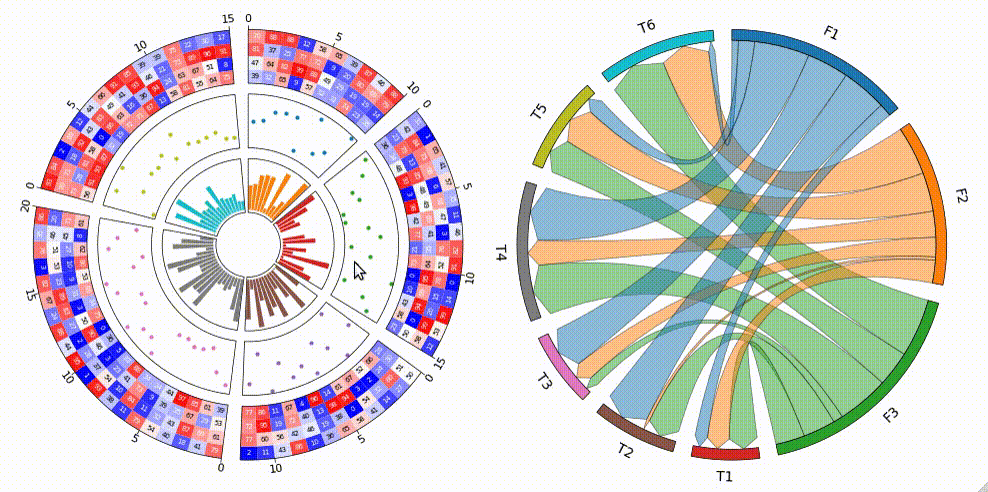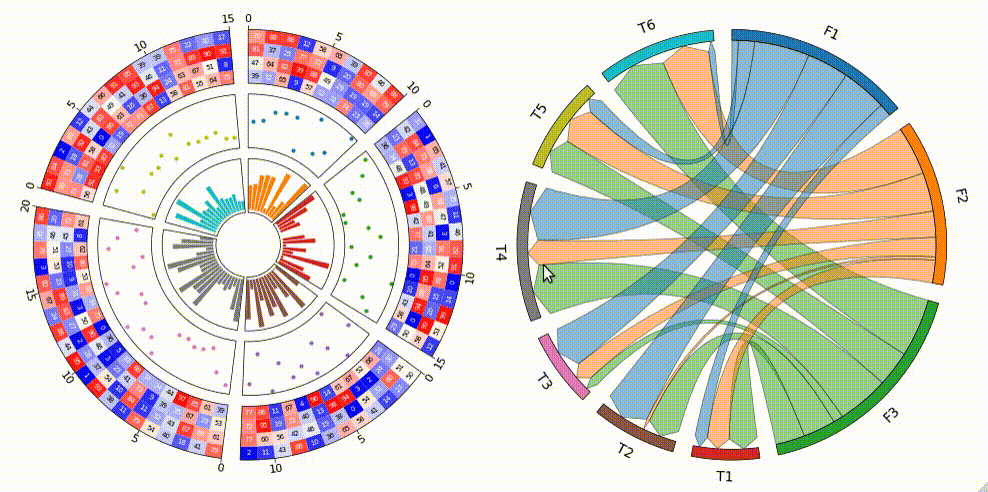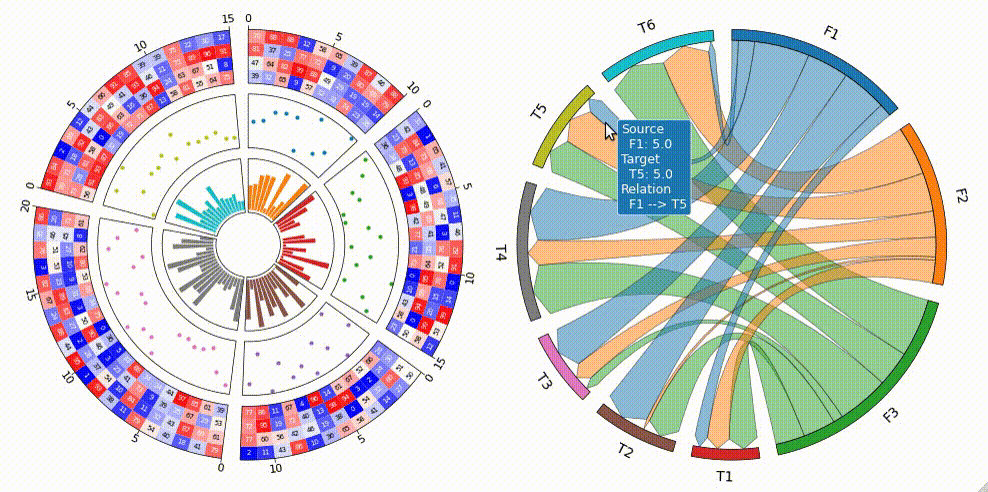
Circular visualization in Python
Project description
pyCirclize: Circular visualization in Python
Table of contents
Overview
pyCirclize is a circular visualization python package implemented based on matplotlib. This package is developed for the purpose of easily and beautifully plotting circular figure such as Circos Plot and Chord Diagram in Python. In addition, useful genome and phylogenetic tree visualization methods for the bioinformatics field are also implemented. pyCirclize was inspired by circlize and pyCircos. More detailed documentation is available here.
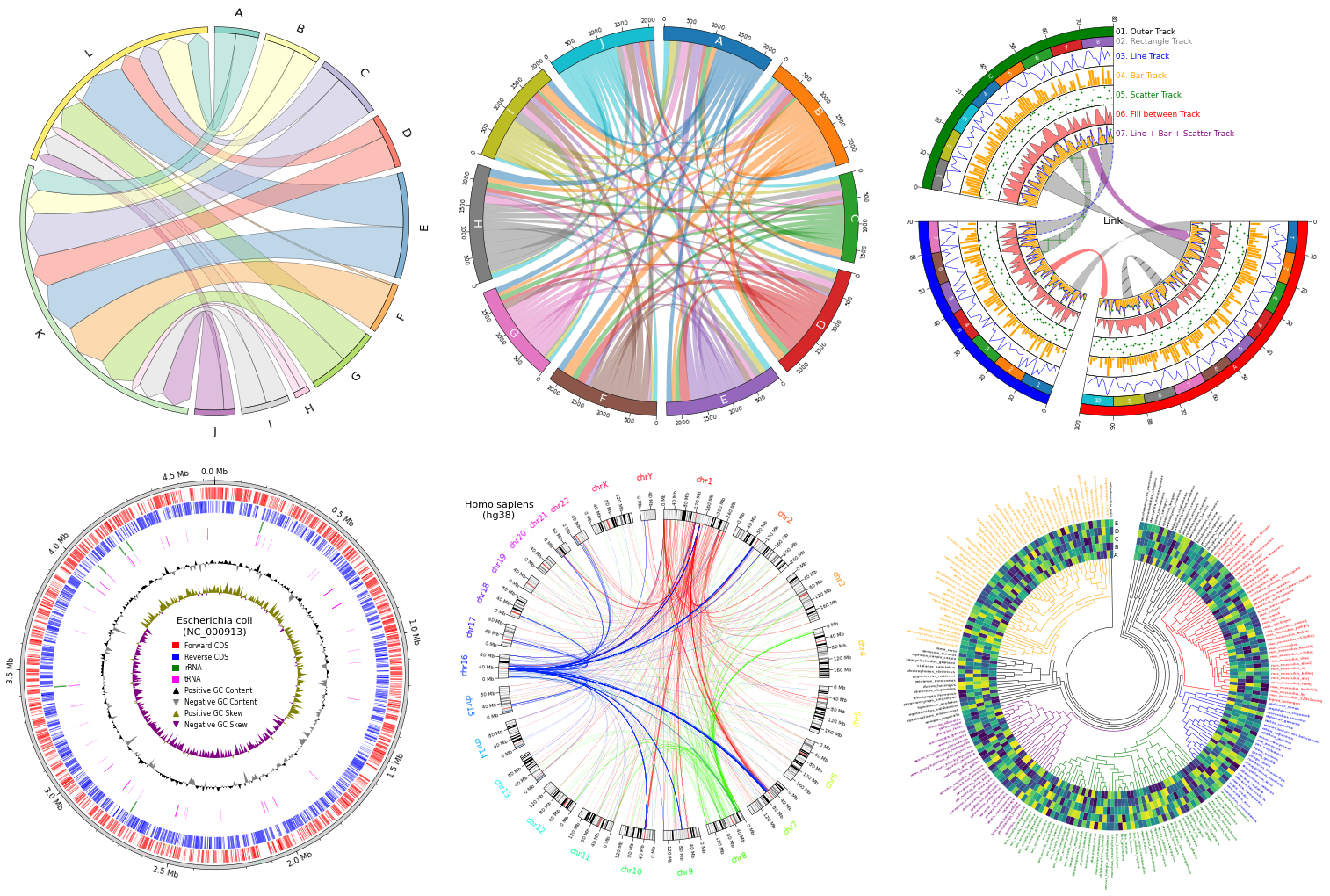
Fig.1 pyCirclize example plot gallery
Installation
Python 3.10 or later is required for installation.
Install PyPI package:
pip install pycirclize
Install conda-forge package:
conda install -c conda-forge pycirclize
API Usage
API usage is described in each of the following sections in the document.
- Getting Started
- Plot API Example
- Chord Diagram
- Radar Chart
- Circos Plot (Genomics)
- Comparative Genomics
- Phylogenetic Tree
- Plot Tips
Code Example
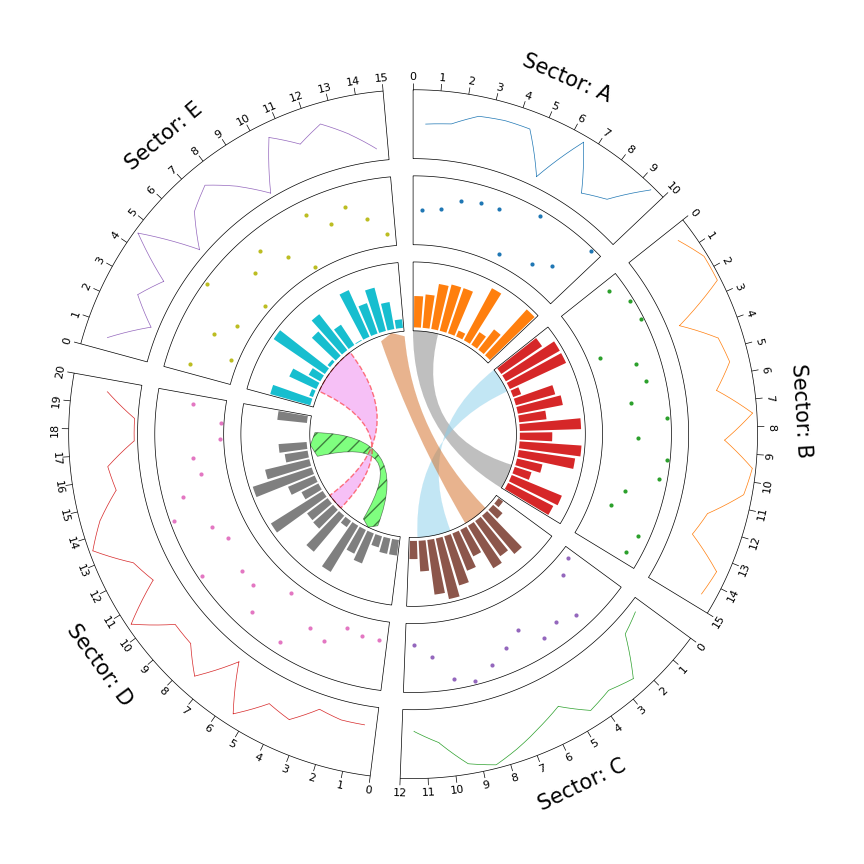
1. Circos Plot
from pycirclize import Circos
import numpy as np
np.random.seed(0)
# Initialize Circos sectors
sectors = {"A": 10, "B": 15, "C": 12, "D": 20, "E": 15}
circos = Circos(sectors, space=5)
for sector in circos.sectors:
# Plot sector name
sector.text(f"Sector: {sector.name}", r=110, size=15)
# Create x positions & random y values
x = np.arange(sector.start, sector.end) + 0.5
y = np.random.randint(0, 100, len(x))
# Plot lines
track1 = sector.add_track((80, 100), r_pad_ratio=0.1)
track1.xticks_by_interval(interval=1)
track1.axis()
track1.line(x, y)
# Plot points
track2 = sector.add_track((55, 75), r_pad_ratio=0.1)
track2.axis()
track2.scatter(x, y)
# Plot bars
track3 = sector.add_track((30, 50), r_pad_ratio=0.1)
track3.axis()
track3.bar(x, y)
# Plot links
circos.link(("A", 0, 3), ("B", 15, 12))
circos.link(("B", 0, 3), ("C", 7, 11), color="skyblue")
circos.link(("C", 2, 5), ("E", 15, 12), color="chocolate", direction=1)
circos.link(("D", 3, 5), ("D", 18, 15), color="lime", ec="black", lw=0.5, hatch="//", direction=2)
circos.link(("D", 8, 10), ("E", 2, 8), color="violet", ec="red", lw=1.0, ls="dashed")
circos.savefig("example01.png")
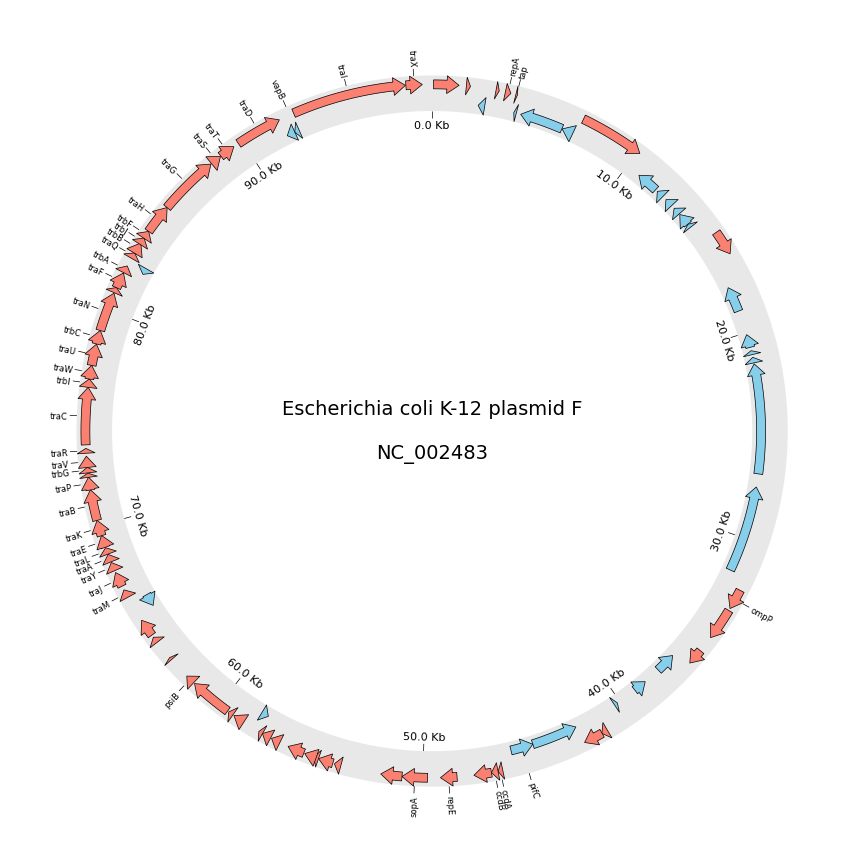
2. Circos Plot (Genomics)
from pycirclize import Circos
from pycirclize.utils import fetch_genbank_by_accid
from pycirclize.parser import Genbank
# Download `NC_002483` E.coli plasmid genbank
gbk_fetch_data = fetch_genbank_by_accid("NC_002483")
gbk = Genbank(gbk_fetch_data)
# Initialize Circos instance with genome size
sectors = gbk.get_seqid2size()
space = 0 if len(sectors) == 1 else 2
circos = Circos(sectors, space=space)
circos.text(f"Escherichia coli K-12 plasmid F\n\n{gbk.name}", size=14)
seqid2features = gbk.get_seqid2features(feature_type="CDS")
for sector in circos.sectors:
# Setup track for features plot
f_cds_track = sector.add_track((95, 100))
f_cds_track.axis(fc="lightgrey", ec="none", alpha=0.5)
r_cds_track = sector.add_track((90, 95))
r_cds_track.axis(fc="lightgrey", ec="none", alpha=0.5)
# Plot forward/reverse strand CDS
features = seqid2features[sector.name]
for feature in features:
if feature.location.strand == 1:
f_cds_track.genomic_features(feature, plotstyle="arrow", fc="salmon", lw=0.5)
else:
r_cds_track.genomic_features(feature, plotstyle="arrow", fc="skyblue", lw=0.5)
# Plot 'gene' qualifier label if exists
labels, label_pos_list = [], []
for feature in features:
start = int(feature.location.start)
end = int(feature.location.end)
label_pos = (start + end) / 2
gene_name = feature.qualifiers.get("gene", [None])[0]
if gene_name is not None:
labels.append(gene_name)
label_pos_list.append(label_pos)
f_cds_track.annotate(label_pos, gene_name, label_size=6)
# Plot xticks (interval = 10 Kb)
r_cds_track.xticks_by_interval(
10000, outer=False, label_formatter=lambda v: f"{v/1000:.1f} Kb"
)
circos.savefig("example02.png")
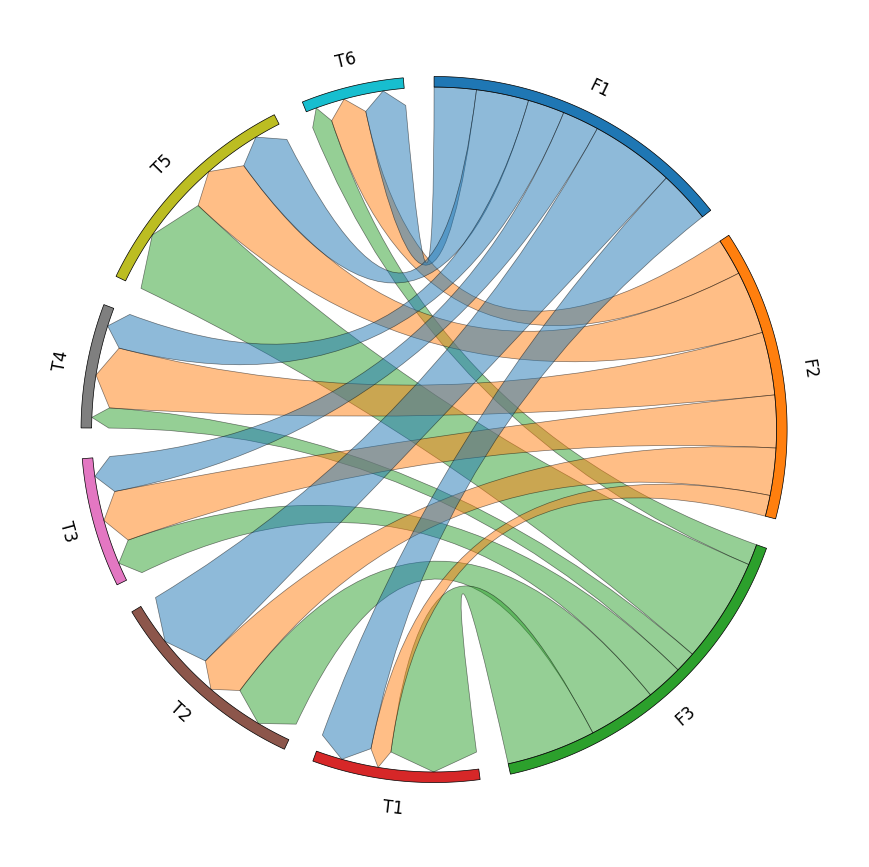
3. Chord Diagram
from pycirclize import Circos
import pandas as pd
# Create matrix dataframe (3 x 6)
row_names = ["F1", "F2", "F3"]
col_names = ["T1", "T2", "T3", "T4", "T5", "T6"]
matrix_data = [
[10, 16, 7, 7, 10, 8],
[4, 9, 10, 12, 12, 7],
[17, 13, 7, 4, 20, 4],
]
matrix_df = pd.DataFrame(matrix_data, index=row_names, columns=col_names)
# Initialize Circos instance for chord diagram plot
circos = Circos.chord_diagram(
matrix_df,
space=5,
cmap="tab10",
label_kws=dict(size=12),
link_kws=dict(ec="black", lw=0.5, direction=1),
)
circos.savefig("example03.png")
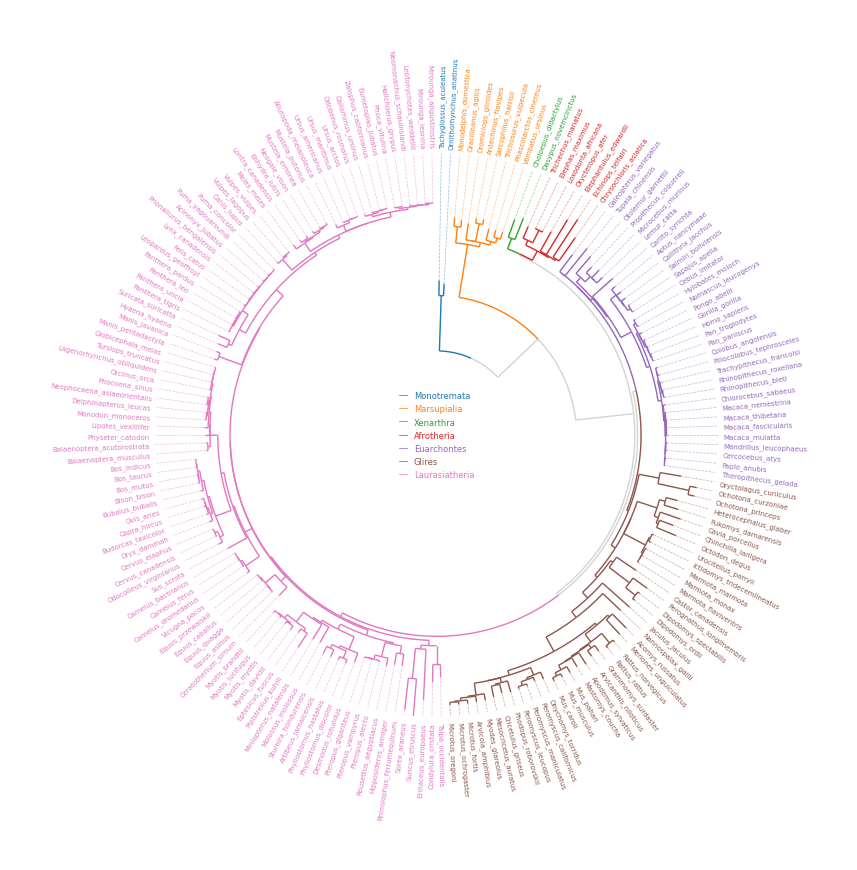
4. Phylogenetic Tree
from pycirclize import Circos
from pycirclize.utils import load_example_tree_file, ColorCycler
from matplotlib.lines import Line2D
# Initialize Circos from phylogenetic tree
tree_file = load_example_tree_file("large_example.nwk")
circos, tv = Circos.initialize_from_tree(
tree_file,
r_lim=(30, 100),
leaf_label_size=5,
line_kws=dict(color="lightgrey", lw=1.0),
)
# Define group-species dict for tree annotation
# In this example, set minimum species list to specify group's MRCA node
group_name2species_list = dict(
Monotremata=["Tachyglossus_aculeatus", "Ornithorhynchus_anatinus"],
Marsupialia=["Monodelphis_domestica", "Vombatus_ursinus"],
Xenarthra=["Choloepus_didactylus", "Dasypus_novemcinctus"],
Afrotheria=["Trichechus_manatus", "Chrysochloris_asiatica"],
Euarchontes=["Galeopterus_variegatus", "Theropithecus_gelada"],
Glires=["Oryctolagus_cuniculus", "Microtus_oregoni"],
Laurasiatheria=["Talpa_occidentalis", "Mirounga_leonina"],
)
# Set tree line color & label color
ColorCycler.set_cmap("tab10")
group_name2color = {name: ColorCycler() for name in group_name2species_list.keys()}
for group_name, species_list in group_name2species_list.items():
color = group_name2color[group_name]
tv.set_node_line_props(species_list, color=color, apply_label_color=True)
# Plot figure & set legend on center
fig = circos.plotfig()
_ = circos.ax.legend(
handles=[Line2D([], [], label=n, color=c) for n, c in group_name2color.items()],
labelcolor=group_name2color.values(),
fontsize=6,
loc="center",
bbox_to_anchor=(0.5, 0.5),
)
fig.savefig("example04.png")
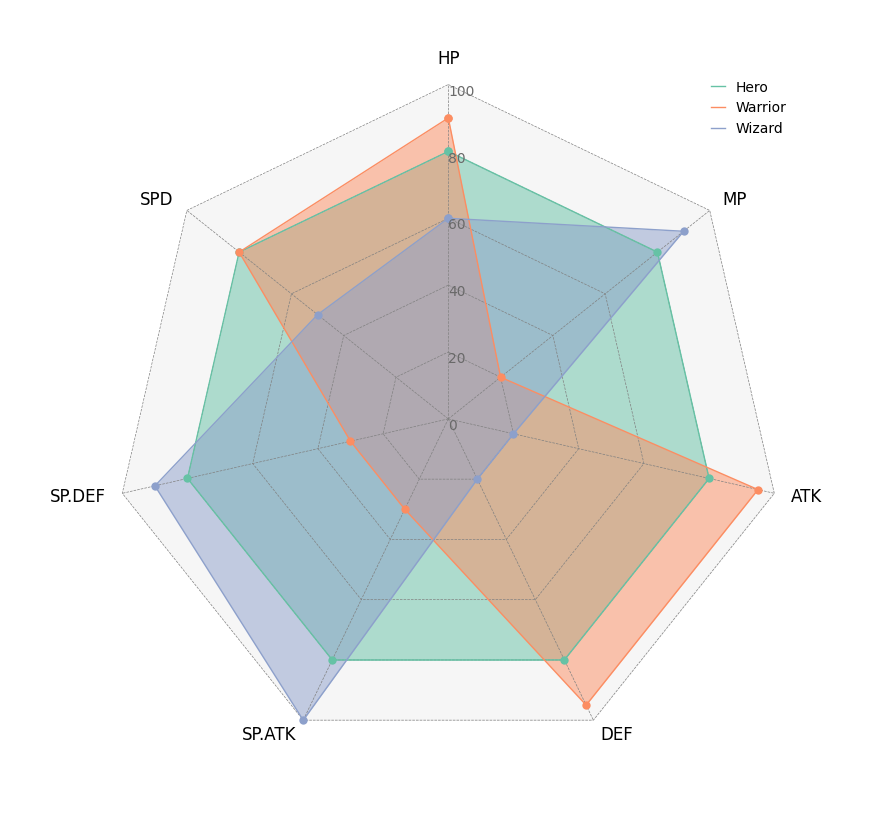
5. Radar Chart
from pycirclize import Circos
import pandas as pd
# Create RPG jobs parameter dataframe (3 jobs, 7 parameters)
df = pd.DataFrame(
data=[
[80, 80, 80, 80, 80, 80, 80],
[90, 20, 95, 95, 30, 30, 80],
[60, 90, 20, 20, 100, 90, 50],
],
index=["Hero", "Warrior", "Wizard"],
columns=["HP", "MP", "ATK", "DEF", "SP.ATK", "SP.DEF", "SPD"],
)
# Initialize Circos instance for radar chart plot
circos = Circos.radar_chart(
df,
vmax=100,
marker_size=6,
grid_interval_ratio=0.2,
)
# Plot figure & set legend on upper right
fig = circos.plotfig()
_ = circos.ax.legend(loc="upper right", fontsize=10)
fig.savefig("example05.png")
Tooltip Option
pyCirclize supports tooltip display in jupyter using ipympl.
To enable tooltip, install pycirclize with ipympl and call circos.plotfig(tooltip=True) method.
Tooltip option is tested on jupyter notebooks in VScode and JupyterLab.
pip install pycirclize[tooltip]
# or
conda install -c conda-forge pycirclize ipympl
[!WARNING] Interactive tooltip plots require live python kernel. Be aware that tooltips are not permanently enabled in the notebook after plotting.
Star History
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pycirclize-1.10.1.tar.gz.
File metadata
- Download URL: pycirclize-1.10.1.tar.gz
- Upload date:
- Size: 20.0 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.8.22
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c67c0bcae01b0c37a07c12f0436540f72c3f1068bfc964f388266ec92812b983
|
|
| MD5 |
24c59c5a31cba0b60f252c7b4576d9d7
|
|
| BLAKE2b-256 |
9f07500623e3db04a4a64d7cf5ab946e5e5e860c198900650a8ea05e1d870d0a
|
File details
Details for the file pycirclize-1.10.1-py3-none-any.whl.
File metadata
- Download URL: pycirclize-1.10.1-py3-none-any.whl
- Upload date:
- Size: 83.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.8.22
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
fc571b0223195b651edd22011963628bddd92861a4053526b31035bb3347f43e
|
|
| MD5 |
58fef9910389e2c3a7a098671960e012
|
|
| BLAKE2b-256 |
35efa0090b20af352b9e8ca7b5ccef81940df63ba15cf5c66baf5c6145620537
|