A set of tools to help building or using Sequana pipelines
Project description






- Overview:
A set of tools to help building or using Sequana pipelines
- Status:
Production
- Issues:
Please fill a report on github
- Python version:
Python 3.8, 3.9, 3.10
- Citation:
Cokelaer et al, (2017), ‘Sequana’: a Set of Snakemake NGS pipelines, Journal of Open Source Software, 2(16), 352, JOSS DOI doi:10.21105/joss.00352
What is sequana_pipetools ?
sequana_pipetools is a collection of tools that assists with the management of Sequana pipelines, which includes next-generation sequencing (NGS) pipelines like RNA-seq, variant calling, ChIP-seq, and others.
The aim of this package is to simplify the deployment of Sequana pipelines by creatin a pure Python library that includse commonly used tools for different pipelines.
Previously, the Sequana framework incorporated alll bioinformatics, Snakemake rules, pipelines, and pipeline management tools into a single library (Sequana) as illustrated in Fig 1 below.
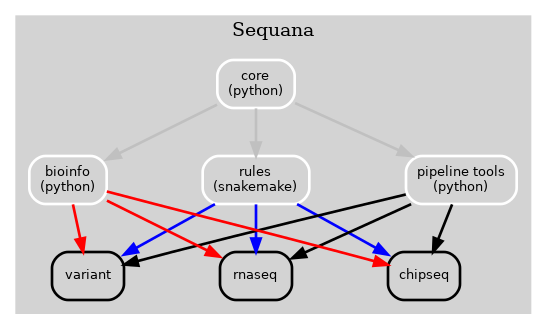
Figure 1 Old Sequana framework will all pipelines and Sequana library in the same place including pipetools (this library).
Whenever changes were made to the Sequana library, a thorough check of the entire library was necessary, despite having 80% test coverage. Adding new pipelines also necessitated the addition of new dependencies, and the process was becoming increasingly complex. To mitigate this issue, we initially made all pipelines independent, as illustrated in Fig. 2. This way, pipeline changes could be made without updating Sequana and vice versa, which was a significant improvment.
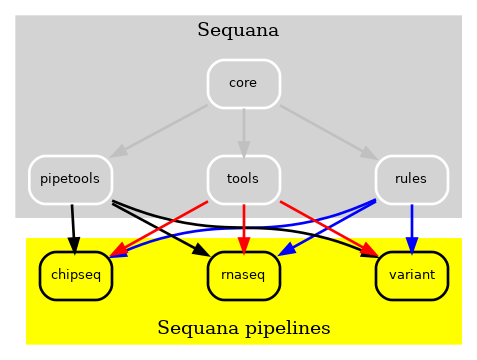
Figure 2 v0.8 of Sequana moved the Snakemake pipelines in indepdendent repositories. A cookie cutter ease the creation of scuh pipelines
However, certain tools, such as those used for user interface and input data sanity checks, were required by all pipelines, as depicted by the pipetools box in the figure. As new pipelines were being added every month, we aimed to make the pipelines and Sequana more modular. Consequently, we created a pure Python library known as sequana_pipetools, as shown in Fig. 3, to make the pipelines even more autonomous.
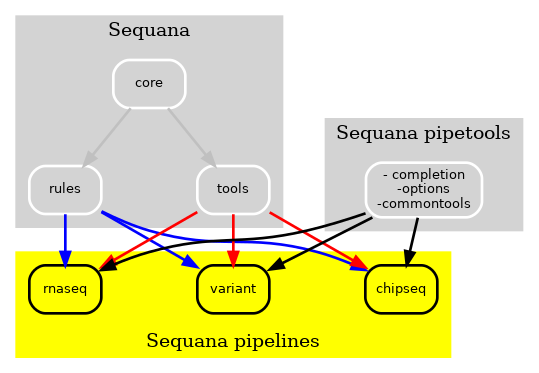
Figure 3 New Sequana framework. The library contains the core and bioinformatics tools and is now distinct from the pipelines. Additionally, the sequana_pipetools library supplies common tools to assist in the creaton and management of all pipelines, such as shared parser for options.
Finally, we dropped the rules/ available in Sequana to build an independent package with a set of Snakemake wrappers. These wrappers are available on https://github.com/sequana/sequana-wrappers and have also the advantage of being tested through continuous integration.
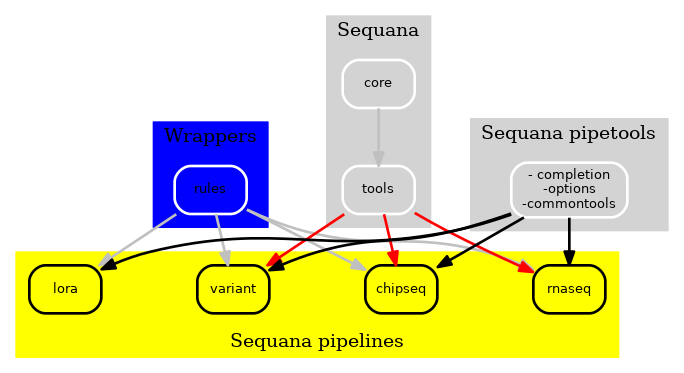
Figure 3 New Sequana framework 2021. The library itself with the core, the bioinformatics tools is now fully independent of the pipelines.
Installation
from pypi website:
pip install sequana_pipetools
No dependencies for this package except Python itself. In practice, this package has no interest if not used with a Sequana pipeline. So, when using it, you will need to install the relevant Sequana pipelines that you wish to use. For example:
pip install sequana_rnaseq pip install sequana_fastqc ...
This package is for Sequana developers. To get more help, go to the doc directory and build the local sphinx directory using:
make html browse build/html/index.html
Quick tour
There are currently two standalone tools. The first one is for Linux users under bash to obtain completion of a sequana pipeline command line arguments:
sequana_completion --name fastqc
The second is used to introspect slurm files to get a summary of the SLURM log files:
sequana_slurm_status --directory .
Will print a short summary report with common errors (if any).
The library is intended to help Sequana developers to design their pipelines. See the Sequana organization repository for examples.
In addition to those standalones, sequana_pipetools goal is to provide utilities to help Sequana developers. We currently provide a set of Options classes that should be used to design the API of your pipelines. For example, the sequana_pipetools.options.SlurmOptions can be used as follows inside a standard Python module (the last two lines is where the magic happens):
import argparse
from sequana_pipetools.options import *
from sequana_pipetools.misc import Colors
from sequana_pipetools.info import sequana_epilog, sequana_prolog
col = Colors()
NAME = "fastqc"
class Options(argparse.ArgumentParser):
def __init__(self, prog=NAME, epilog=None):
usage = col.purple(sequana_prolog.format(**{"name": NAME}))
super(Options, self).__init__(usage=usage, prog=prog, description="",
epilog=epilog,
formatter_class=argparse.ArgumentDefaultsHelpFormatter
)
# add a new group of options to the parser
so = SlurmOptions()
so.add_options(self)
Developers should look at e.g. module sequana_pipetools.options for the API reference and one of the official sequana pipeline (e.g., https://github.com/sequana/sequana_variant_calling) to get help from examples.
The Options classes provided can be used and combined to design pipelines.
Setting up and Running Sequana pipelines
When you execute a sequana pipeline, e.g.:
sequana_fastqc --input-directory data
a working directory is created (with the name of the pipeline; here fastqc). Moreover, the working directory contains a shell script that will hide the snakemake command. This snakemake command with make use of the sequana wrappers and will use the official sequana github repository by default (https://github.com/sequana/sequana-wrappers). This may be overwritten. For instance, you may use a local clone. To do so, you will need to create an environment variable:
export SEQUANA_WRAPPERS="git+file:///home/user/github/sequana-wrappers
If you decide to use singularity/apptainer, one common error on a cluster is that non-standard paths are not found. You can bind them using the -B option but a more general set up is to create thos environment variable:
export SINGULARITY_BINDPATH=" /path_to_bind"
for Singularity setup, or
export APPTAINER_BINDPATH=" /path_to_bind"
for Apptainer setup.
What is Sequana ?
Sequana is a versatile tool that provides
A Python library dedicated to NGS analysis (e.g., tools to visualise standard NGS formats).
A set of Pipelines dedicated to NGS in the form of Snakefiles (Makefile-like with Python syntax based on snakemake framework) with more than 80 re-usable rules.
Standalone applications.
See the sequana home page for details.
To join the project, please let us know on github.
Changelog
Version |
Description |
|---|---|
0.12.0 |
|
0.11.1 |
|
0.11.0 |
|
0.10.2 |
|
0.10.1 |
|
0.10.0 |
|
0.9.6 |
|
0.9.5 |
|
0.9.4 |
|
0.9.3 |
|
0.9.2 |
|
0.9.1 |
|
0.9.0 |
|
0.8.1 |
|
0.8.0 |
|
0.7.6 |
|
0.7.5 |
|
0.7.4 |
|
0.7.3 |
|
0.7.2 |
|
0.7.1 |
|
0.7.0 |
|
0.6.3 |
|
0.6.2 |
|
0.6.1 |
|
0.6.0 |
|
0.5.3 |
|
0.5.2 |
|
0.5.1 |
|
0.5.0 |
|
0.4.3 |
|
0.4.2 |
|
0.4.1 |
|
0.4.0 |
|
0.3.1 |
|
0.3.0 |
|
0.2.6 |
|
0.2.5 |
|
0.2.4 |
|
0.2.3 |
|
0.2.2 |
|
0.2.1 |
|
0.2.0 |
add content from sequana.pipeline_common to handle all kind of options in the argparse of all pipelines. This is independent of sequana to speed up the –version and –help calls |
0.1.2 |
add version of the pipeline in the output completion file |
0.1.1 |
release bug fix |
0.1.0 |
creation of the package |
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.











