Small library for creating and visualizing dot plot matrices
Project description
wotplot
wotplot is a small Python library for creating and visualizing dot plot matrices.
Quick examples
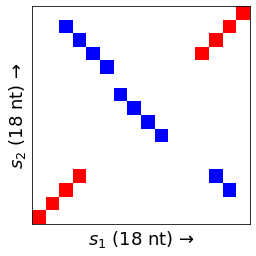
Small dataset
This example is adapted from Figure 6.20 (bottom right) in Bioinformatics Algorithms, edition 2.
import wotplot
# Define our dataset
s1 = "AGCAGGAGATAAACCTGT"
s2 = "AGCAGGTTATCTACCTGT"
k = 3
# Create the matrix (the "binary" parameter means we'll distinguish forward,
# reverse-complementary, and palindromic matching k-mers from each other)
m = wotplot.DotPlotMatrix(s1, s2, k, binary=False)
# Convert the matrix to dense format and visualize it using matplotlib's
# imshow() function (for large matrices where dense representations are
# impractical, use viz_spy() instead; see below)
wotplot.viz_imshow(m)
In the default colorscheme red cells (🟥) indicate forward matches, blue cells (🟦) indicate reverse-complementary matches, and purple cells (🟪) indicate palindromic matches.
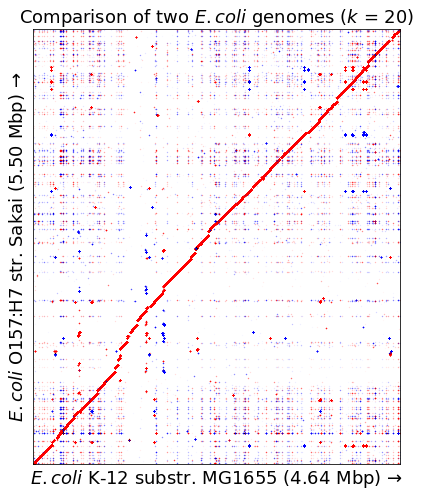
Larger dataset: comparing two E. coli genomes
Using E. coli K-12 (from this assembly) and E. coli O157:H7 (from this assembly). I removed the two plasmid sequences from the O157:H7 assembly.
import wotplot
from matplotlib import pyplot
# (skipping the part where I loaded the genomes into memory as e1s and e2s...)
# Create the matrix (leaving binary=True by default)
em = wotplot.DotPlotMatrix(e1s, e2s, 20, verbose=True)
# Visualize the matrix using matplotlib's spy() function
fig, ax = pyplot.subplots()
wotplot.viz_spy(
em, markersize=0.01, title="Comparison of two $E. coli$ genomes ($k$ = 20)", ax=ax
)
ax.set_xlabel(f"$E. coli$ K-12 substr. MG1655 ({len(e1s)/1e6:.2f} Mbp) \u2192")
ax.set_ylabel(f"$E. coli$ O157:H7 str. Sakai ({len(e2s)/1e6:.2f} Mbp) \u2192")
fig.set_size_inches(8, 8)
More detailed tutorial
Please see this Jupyter Notebook.
Installation
wotplot supports Python ≥ 3.6. You can install it and its dependencies using pip:
pip install git+https://github.com/fedarko/wotplot.git
I'll try to put this on PyPI / conda eventually.
Performance
Optimizations made so far
I've tried to make this library reasonably performant. The main optimizations include:
-
We use suffix arrays (courtesy of the lovely
pydivsufsortlibrary) in order to reduce the memory footprint of finding shared k-mers. -
We store the dot plot matrix in sparse format (courtesy of SciPy) in order to reduce its memory footprint.
-
We support visualizing the dot plot matrix's nonzero values using matplotlib's
spy()function, which (at least for large sequences) is faster and more memory-efficient than converting the matrix to a dense format and visualizing it with something likeimshow().
That being said...
This library could be made a lot more efficient (I've been documenting ideas in issue #2), but right now it's good enough for my purposes. Feel free to open an issue / make a pull request if you'd like to speed it up ;)
Informal benchmarking
See this Jupyter Notebook for some very informal benchmarking results performed on a laptop with ~8 GB of RAM.
Even on this system, the library can handle reasonably large sequences: in the biggest example, the notebook demonstrates computing the dot plot of two random 100 Mbp sequences (using k = 20) in 54 minutes and 12.45 seconds. Dot plots of shorter sequences (e.g. 100 kbp or less) usually take only a few seconds to compute, at least for reasonably large values of k.
Why does this library exist?
-
This library separates the creation and visualization of dot plot matrices. Other tools that I tried produced pretty visualizations, but didn't give me easy access to the underlying matrix.
-
I wanted something that worked well with matplotlib, so that I could create and tile lots of dotplots at once in complicated ways.
Setting up a development environment
First, fork wotplot. Then you can download a copy of the code from your fork and install wotplot from this code.
The following commands should work on a Unix system; this assumes that you have Python ≥ 3.6 and pip installed.
git clone https://github.com/your-github-username-goes-here/wotplot.git
cd wotplot
pip install -e .[dev]
Contact
Feel free to open an issue if you have questions, suggestions, comments, or anything else.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file wotplot-0.1.0.tar.gz.
File metadata
- Download URL: wotplot-0.1.0.tar.gz
- Upload date:
- Size: 14.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.8.0 pkginfo/1.9.6 readme-renderer/34.0 requests/2.27.1 requests-toolbelt/1.0.0 urllib3/1.26.16 tqdm/4.64.1 importlib-metadata/4.2.0 keyring/23.4.1 rfc3986/1.5.0 colorama/0.4.5 CPython/3.6.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
645763a66e902074c216921b258eebfbedcdd6ea297c339c2e87904843d01d91
|
|
| MD5 |
d5d9aa7b4e103c1a41f14f4e1f5d4359
|
|
| BLAKE2b-256 |
2cf0276b6b99ffdc9db043a34b1d381777127b291b39af6c02c222dabc6b69ac
|
File details
Details for the file wotplot-0.1.0-py3-none-any.whl.
File metadata
- Download URL: wotplot-0.1.0-py3-none-any.whl
- Upload date:
- Size: 15.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.8.0 pkginfo/1.9.6 readme-renderer/34.0 requests/2.27.1 requests-toolbelt/1.0.0 urllib3/1.26.16 tqdm/4.64.1 importlib-metadata/4.2.0 keyring/23.4.1 rfc3986/1.5.0 colorama/0.4.5 CPython/3.6.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9652c914141d358a3e9a85a861af9a0339882c159c845f650254d43b02c587eb
|
|
| MD5 |
04dd5dc3a2aa1a0bae79d47c77d3f482
|
|
| BLAKE2b-256 |
6a83b79573063711fc1f79f0b5920f7a210c6645117d457d462a0f24f8dcdafe
|