A package for decoding quantum error correcting codes using minimum-weight perfect matching.
Project description
PyMatching 2
PyMatching is a fast Python/C++ library for decoding quantum error correcting (QEC) codes using the Minimum Weight Perfect Matching (MWPM) decoder. Given the syndrome measurements from a quantum error correction circuit, the MWPM decoder finds the most probable set of errors, given the assumption that error mechanisms are independent, as well as graphlike (each error causes either one or two detection events). The MWPM decoder is the most popular decoder for decoding surface codes, and can also be used to decode various other code families, including subsystem codes, honeycomb codes and 2D hyperbolic codes.
Version 2 includes a new implementation of the blossom algorithm which is 100-1000x faster than previous versions of PyMatching. PyMatching can be configured using arbitrary weighted graphs, with or without a boundary, and can be combined with Craig Gidney's Stim library to simulate and decode error correction circuits in the presence of circuit-level noise. The sinter package combines Stim and PyMatching to perform fast, parallelised monte-carlo sampling of quantum error correction circuits. As of a recent update (v2.3), pymatching also supports correlated matching.
Documentation for PyMatching can be found at: pymatching.readthedocs.io
Our paper gives more background on the MWPM decoder and our implementation (sparse blossom) released in PyMatching v2.
To see how stim, sinter and pymatching can be used to estimate the threshold of an error correcting code with circuit-level noise, try out the stim getting started notebook.
The new >100x faster implementation for Version 2
Version 2 features a new implementation of the blossom algorithm, which I wrote with Craig Gidney. Our new implementation, which we refer to as the sparse blossom algorithm, can be seen as a generalisation of the blossom algorithm to handle the decoding problem relevant to QEC. We solve the problem of finding minimum-weight paths between detection events in a detector graph directly, which avoids the need to use costly all-to-all Dijkstra searches to find a MWPM in a derived graph using the original blossom algorithm. The new version is also exact - unlike previous versions of PyMatching, no approximation is made. See our paper for more details.
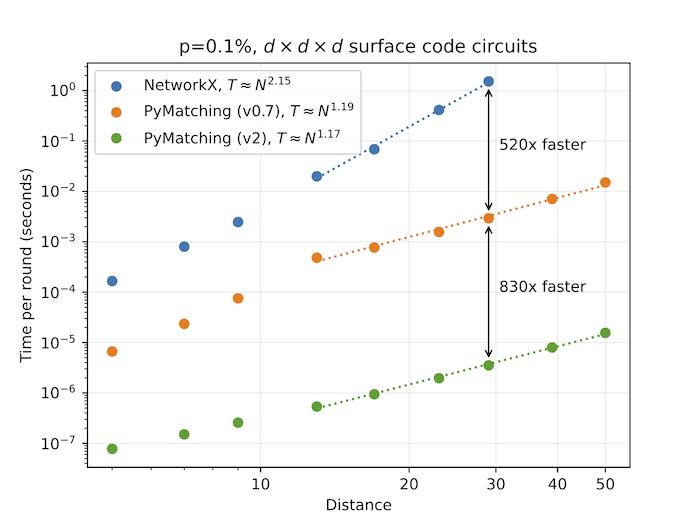
Our new implementation is over 100x faster than previous versions of PyMatching, and is over 100,000x faster than NetworkX (benchmarked with surface code circuits). At 0.1% circuit-noise, PyMatching can decode both X and Z basis measurements of surface code circuits up to distance 17 in under 1 microsecond per round of syndrome extraction on a single core. Furthermore, the runtime is roughly linear in the number of nodes in the graph.
The plot below compares the performance of PyMatching v2 with the previous version (v0.7) as well as with NetworkX for decoding surface code circuits with circuit-level depolarising noise. All decoders were run on a single core of an M1 processor, processing both the X and Z basis measurements. The equations T=N^x in the legend (and plotted as dashed lines) are obtained from a fit to the same dataset for distance > 10, where N is the number of detectors (nodes) per round, and T is the decoding time per round. See the benchmarks folder in the repository for the data and stim circuits, as well as additional benchmarks.
Sparse blossom is conceptually similar to the approach described in this paper by Austin Fowler, although our approach differs in many of the details (as explained in our paper). There are even more similarities with the very nice independent work by Yue Wu, who recently released the fusion-blossom library. One of the differences with our approach is that fusion-blossom grows the exploratory regions of alternating trees in a similar way to how clusters are grown in Union-Find, whereas our approach instead progresses along a timeline, and uses a global priority queue to grow alternating trees. Yue also has a paper coming soon, so stay tuned for that as well.
Correlated matching
As of PyMatching version 2.3, correlated matching is now also available in pymatching! Thank you to Sid Madhuk, who was the primary contributor for this new feature.
Correlated matching has better accuracy than standard (uncorrelated) matching for many decoding problems where hyperedge errors are present. When these hyperedge errors are decomposed into edges (graphlike errors), they amount to correlations between these edges in the matching graph. A common example of such a hyperedge error is a $Y$ error in the surface code.
The "two-pass" correlated matching decoder implemented in pymatching works by running sparse blossom twice. The first pass is a standard (uncorrelated) run of sparse blossom, to predict a set of edges in the matching graph. Correlated matching then assumes these errors (edges) occurred and reweights edges that are correlated with it based on this assumption. Matching is then run for the second time on this reweighted graph.
Installation
The latest version of PyMatching can be downloaded and installed from PyPI with the command:
pip install pymatching --upgrade
Usage
PyMatching can load matching graphs from a check matrix, a stim.DetectorErrorModel, a networkx.Graph, a
rustworkx.PyGraph or by adding edges individually with pymatching.Matching.add_edge and
pymatching.Matching.add_boundary_edge.
Decoding Stim circuits
PyMatching can be combined with Stim. Generally, the easiest and fastest way to
do this is using sinter (use v1.10.0 or later), which uses PyMatching and Stim to run
parallelised monte carlo simulations of quantum error correction circuits.
However, in this section we will use Stim and PyMatching directly, to demonstrate how their Python APIs can be used.
To install stim, run pip install stim --upgrade.
First, we generate a stim circuit. Here, we use a surface code circuit included with stim:
import numpy as np
import stim
import pymatching
circuit = stim.Circuit.generated(
"surface_code:rotated_memory_x",
distance=5,
rounds=5,
after_clifford_depolarization=0.005
)
Next, we use stim to generate a stim.DetectorErrorModel (DEM), which is effectively a
Tanner graph describing the circuit-level noise model.
By setting decompose_errors=True, stim decomposes all error mechanisms into edge-like error
mechanisms (which cause either one or two detection events).
This ensures that our DEM is graphlike, and can be loaded by pymatching:
model = circuit.detector_error_model(decompose_errors=True)
matching = pymatching.Matching.from_detector_error_model(model)
Next, we will sample 1000 shots from the circuit. Each shot (a row of shots) contains the full syndrome (detector
measurements), as well as the logical observable measurements, from simulating the noisy circuit:
sampler = circuit.compile_detector_sampler()
syndrome, actual_observables = sampler.sample(shots=1000, separate_observables=True)
Now we can decode! We compare PyMatching's predictions of the logical observables with the actual observables sampled with stim, in order to count the number of mistakes and estimate the logical error rate:
predicted_observables = matching.decode_batch(syndrome)
num_errors = np.sum(np.any(predicted_observables != actual_observables, axis=1))
print(num_errors) # prints 8
To decode instead with correlated matching, set enable_correlations=True both when configuiing the pymatching.Matching object:
matching_corr = pymatching.Matching.from_detector_error_model(model, enable_correlations=True)
as well as when decoding:
predicted_observables_corr = matching_corr.decode_batch(syndrome, enable_correlations=True)
num_errors = np.sum(np.any(predicted_observables_corr != actual_observables, axis=1))
print(num_errors) # prints 3
Loading from a parity check matrix
We can also load a pymatching.Matching object from a binary
parity check matrix, another representation of a Tanner graph.
Each row in the parity check matrix H corresponds to a parity check, and each column corresponds to an
error mechanism.
The element H[i,j] of H is 1 if parity check i is flipped by error mechanism j, and 0 otherwise.
To be used by PyMatching, the error mechanisms in H must be graphlike.
This means that each column must contain either one or two 1s (if a column has a single 1, it represents a half-edge
connected to the boundary).
We can give each edge in the graph a weight, by providing PyMatching with a weights numpy array.
Element weights[j] of the weights array sets the edge weight for the edge corresponding to column j of H.
If the error mechanisms are treated as independent, then we typically want to set the weight of edge j to
the log-likelihood ratio log((1-p_j)/p_j), where p_j is the error probability associated with edge j.
With this setting, PyMatching will find the most probable set of error mechanisms, given the syndrome.
With PyMatching configured using H and weights, decoding a binary syndrome vector syndrome (a numpy array
of length H.shape[0]) corresponds to finding a set of errors defined in a binary predictions vector
satisfying H@predictions % 2 == syndrome while minimising the total solution weight predictions@weights.
In quantum error correction, rather than predicting which exact set of error mechanisms occurred, we typically want to
predict the outcome of logical observable measurements, which are the parities of error mechanisms.
These can be represented by a binary matrix observables. Similar to the check matrix, observables[i,j] is 1 if
logical observable i is flipped by error mechanism j.
For example, suppose our syndrome syndrome, was the result of a set of errors noise (a binary array of
length H.shape[1]), such that syndrome = H@noise % 2.
Our decoding is successful if observables@noise % 2 == observables@predictions % 2.
Putting this together, we can decode a distance 5 repetition code as follows:
import numpy as np
from scipy.sparse import csc_matrix
import pymatching
H = csc_matrix([[1, 1, 0, 0, 0],
[0, 1, 1, 0, 0],
[0, 0, 1, 1, 0],
[0, 0, 0, 1, 1]])
weights = np.array([4, 3, 2, 3, 4]) # Set arbitrary weights for illustration
matching = pymatching.Matching(H, weights=weights)
prediction = matching.decode(np.array([0, 1, 0, 1]))
print(prediction) # prints: [0 0 1 1 0]
# Optionally, we can return the weight as well:
prediction, solution_weight = matching.decode(np.array([0, 1, 0, 1]), return_weight=True)
print(prediction) # prints: [0 0 1 1 0]
print(solution_weight) # prints: 5.0
And in order to estimate the logical error rate for a physical error rate of 10%, we can sample as follows:
import numpy as np
from scipy.sparse import csc_matrix
import pymatching
H = csc_matrix([[1, 1, 0, 0, 0],
[0, 1, 1, 0, 0],
[0, 0, 1, 1, 0],
[0, 0, 0, 1, 1]])
observables = csc_matrix([[1, 0, 0, 0, 0]])
error_probability = 0.1
weights = np.ones(H.shape[1]) * np.log((1-error_probability)/error_probability)
matching = pymatching.Matching.from_check_matrix(H, weights=weights)
num_shots = 1000
num_errors = 0
for i in range(num_shots):
noise = (np.random.random(H.shape[1]) < error_probability).astype(np.uint8)
syndrome = H@noise % 2
prediction = matching.decode(syndrome)
predicted_observables = observables@prediction % 2
actual_observables = observables@noise % 2
num_errors += not np.array_equal(predicted_observables, actual_observables)
print(num_errors) # prints 4
Note that we can also ask PyMatching to predict the logical observables directly, by supplying them
to the faults_matrix argument when constructing the pymatching.Matching object. This allows the decoder to make
some additional optimisations, that speed up the decoding procedure a bit. The following example uses this approach,
and is equivalent to the example above:
import numpy as np
from scipy.sparse import csc_matrix
import pymatching
H = csc_matrix([[1, 1, 0, 0, 0],
[0, 1, 1, 0, 0],
[0, 0, 1, 1, 0],
[0, 0, 0, 1, 1]])
observables = csc_matrix([[1, 0, 0, 0, 0]])
error_probability = 0.1
weights = np.ones(H.shape[1]) * np.log((1-error_probability)/error_probability)
matching = pymatching.Matching.from_check_matrix(H, weights=weights, faults_matrix=observables)
num_shots = 1000
num_errors = 0
for i in range(num_shots):
noise = (np.random.random(H.shape[1]) < error_probability).astype(np.uint8)
syndrome = H@noise % 2
predicted_observables = matching.decode(syndrome)
actual_observables = observables@noise % 2
num_errors += not np.array_equal(predicted_observables, actual_observables)
print(num_errors) # prints 6
We'll make one more optimisation, which is to use matching.decode_batch to decode the batch of shots, rather than
iterating over calls to matching.decode in Python:
import numpy as np
from scipy.sparse import csc_matrix
import pymatching
H = csc_matrix([[1, 1, 0, 0, 0],
[0, 1, 1, 0, 0],
[0, 0, 1, 1, 0],
[0, 0, 0, 1, 1]])
observables = csc_matrix([[1, 0, 0, 0, 0]])
error_probability = 0.1
num_shots = 1000
weights = np.ones(H.shape[1]) * np.log((1-error_probability)/error_probability)
matching = pymatching.Matching.from_check_matrix(H, weights=weights, faults_matrix=observables)
noise = (np.random.random((num_shots, H.shape[1])) < error_probability).astype(np.uint8)
shots = (noise @ H.T) % 2
actual_observables = (noise @ observables.T) % 2
predicted_observables = matching.decode_batch(shots)
num_errors = np.sum(np.any(predicted_observables != actual_observables, axis=1))
print(num_errors) # prints 6
Instead of using a check matrix, the Matching object can also be constructed using
the Matching.add_edge
and
Matching.add_boundary_edge
methods, or by loading from a NetworkX or rustworkx graph.
For more details on how to use PyMatching, see the documentation.
Attribution
When using PyMatching please cite our paper on the sparse blossom algorithm (implemented in version 2):
@article{Higgott2025sparseblossom,
doi = {10.22331/q-2025-01-20-1600},
url = {https://doi.org/10.22331/q-2025-01-20-1600},
title = {Sparse {B}lossom: correcting a million errors per core second with minimum-weight matching},
author = {Higgott, Oscar and Gidney, Craig},
journal = {{Quantum}},
issn = {2521-327X},
publisher = {{Verein zur F{\"{o}}rderung des Open Access Publizierens in den Quantenwissenschaften}},
volume = {9},
pages = {1600},
month = jan,
year = {2025}
}
Note: the previous PyMatching paper descibes the implementation in version 0.7 and earlier of PyMatching (not v2).
Acknowledgements
We are grateful to the Google Quantum AI team for supporting the development of PyMatching v2. Earlier versions of PyMatching were supported by Unitary Fund and EPSRC.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distributions
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pymatching-2.3.1.tar.gz.
File metadata
- Download URL: pymatching-2.3.1.tar.gz
- Upload date:
- Size: 347.1 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
dac1afab0a190a003a3689a18c3afc5c99a5beaa6a099d79e3d4c8b24c1fa7c5
|
|
| MD5 |
aecc898dfe1e0f9140e06cb126576e5e
|
|
| BLAKE2b-256 |
c59260f1419500b49afb0925b0536538c352ef71b4bf182c28b0e3861141fc81
|
File details
Details for the file pymatching-2.3.1-cp313-cp313-win_amd64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp313-cp313-win_amd64.whl
- Upload date:
- Size: 348.0 kB
- Tags: CPython 3.13, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
7ebce2e8bc98379f0bb7643a0ef0171142cf384ba626e46108c84249c868f0a6
|
|
| MD5 |
91690ec38fcb635b9d44663c8eacb6c2
|
|
| BLAKE2b-256 |
1fc40e53ff3c9312ffb0e1677ebb62172660c5614077dca9c69ccfc31b90b7e4
|
File details
Details for the file pymatching-2.3.1-cp313-cp313-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp313-cp313-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl
- Upload date:
- Size: 625.9 kB
- Tags: CPython 3.13, manylinux: glibc 2.27+ x86-64, manylinux: glibc 2.28+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
d0edddbae8437d173d7c1314c9cb853d4abc9b84be9b2274f76972ba89b1b5b5
|
|
| MD5 |
1bc6d6098083cdea7391b748480255ee
|
|
| BLAKE2b-256 |
6f220d0494562792575be67358dcd8951678449b273af168dd5e52b887e51ab1
|
File details
Details for the file pymatching-2.3.1-cp313-cp313-macosx_11_0_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp313-cp313-macosx_11_0_x86_64.whl
- Upload date:
- Size: 413.0 kB
- Tags: CPython 3.13, macOS 11.0+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
8e3122358a398853d9e809beadcf52b2d7f38a7cac21e14bca6ce0b44045c376
|
|
| MD5 |
ffab70267d568d146cf4c969ed219f92
|
|
| BLAKE2b-256 |
f8efa3114a4c29cd43e180f0c494aa71beda969a347ee34459c4f816597d5e53
|
File details
Details for the file pymatching-2.3.1-cp313-cp313-macosx_11_0_arm64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp313-cp313-macosx_11_0_arm64.whl
- Upload date:
- Size: 380.3 kB
- Tags: CPython 3.13, macOS 11.0+ ARM64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5c010d2dc808a58e80874ada568a4d2bb8c9e81959b858e2e56a84ccc6b2b4ac
|
|
| MD5 |
20df754c347b43def3e3c63198ba0aa6
|
|
| BLAKE2b-256 |
e0ee1e89ba6a2811fad9f1c8d4254812c97904ee386e0490cdd0ee8c68d5d1cd
|
File details
Details for the file pymatching-2.3.1-cp312-cp312-win_amd64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp312-cp312-win_amd64.whl
- Upload date:
- Size: 347.9 kB
- Tags: CPython 3.12, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
1f3a0d307b30ef52b7a9cabb58ec27a08ef057e09bcbd00ffe3c40f4e31e606d
|
|
| MD5 |
0f2304acdbce693239951b3f1136501f
|
|
| BLAKE2b-256 |
d950e34731b6e7f1a46facaa5c189cc95a55b49a00730203d09f211ef4d5ee58
|
File details
Details for the file pymatching-2.3.1-cp312-cp312-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp312-cp312-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl
- Upload date:
- Size: 626.1 kB
- Tags: CPython 3.12, manylinux: glibc 2.27+ x86-64, manylinux: glibc 2.28+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b90cf168645adb78c9f81dd60c9e97e2aae005386b5795b54c8b4055d1492b1b
|
|
| MD5 |
bd6584c97a61fe21a1574c73359173e5
|
|
| BLAKE2b-256 |
dab525e97b533495a839780c89dac2b56e2b683100945ac4149645f9d95e7aee
|
File details
Details for the file pymatching-2.3.1-cp312-cp312-macosx_11_0_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp312-cp312-macosx_11_0_x86_64.whl
- Upload date:
- Size: 412.9 kB
- Tags: CPython 3.12, macOS 11.0+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
fb06a9a60c32c490f2637a5d0ccb70c8c53024f970ab50d699bc8dc1c61f0534
|
|
| MD5 |
3f6663f54065e63aed5fb79308bac1ef
|
|
| BLAKE2b-256 |
756090c3fd4fe1b16dfc46aa3a774c9dc0438299aa07e7d9f41c3f6bbd207734
|
File details
Details for the file pymatching-2.3.1-cp312-cp312-macosx_11_0_arm64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp312-cp312-macosx_11_0_arm64.whl
- Upload date:
- Size: 380.2 kB
- Tags: CPython 3.12, macOS 11.0+ ARM64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
4bde7bac71e3b1b1d55569c664a7a2d0b0d87962df12b9b45ce3e287d0ac0dff
|
|
| MD5 |
75cd24162f7f525877c8fe7842d835e0
|
|
| BLAKE2b-256 |
33e9cf2302b391f7c692feed5c1cddb07c2667722dd2944232cfb4c1a691e2ca
|
File details
Details for the file pymatching-2.3.1-cp311-cp311-win_amd64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp311-cp311-win_amd64.whl
- Upload date:
- Size: 346.0 kB
- Tags: CPython 3.11, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2d27bd5cf79b25268dfce11fb9e3a47acd0a5c1f88815c2fab333eaa8bd45b7b
|
|
| MD5 |
537bf4f1e0c7e0bcc605c4a31dfe8196
|
|
| BLAKE2b-256 |
569714767cb0cb81de1cd2fc3211072bc3bdd08d0d4ae054eafa5584273a6a53
|
File details
Details for the file pymatching-2.3.1-cp311-cp311-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp311-cp311-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl
- Upload date:
- Size: 624.6 kB
- Tags: CPython 3.11, manylinux: glibc 2.27+ x86-64, manylinux: glibc 2.28+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
d462371c7d9864fb1b7993018f16e2f105d732e5671c56dd14c9a789a1aa4861
|
|
| MD5 |
44b75aa13490ba1640bd469eabf8562f
|
|
| BLAKE2b-256 |
24dc946e33cdc7aa734913f2c97326ddcea0f0fdd3cfdcd7b986e5326f5d3ee3
|
File details
Details for the file pymatching-2.3.1-cp311-cp311-macosx_11_0_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp311-cp311-macosx_11_0_x86_64.whl
- Upload date:
- Size: 412.9 kB
- Tags: CPython 3.11, macOS 11.0+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
10699180d84ec27a6bf5ccd78c61b8aa52afff0f01263cf928a51ec876138037
|
|
| MD5 |
cfa37f3b5dcf76b8e04c904d155e094b
|
|
| BLAKE2b-256 |
4224a72c64d30b7f1a86b3b87bd358b01c00eb2a8da8ceda9d3e4b030f73f26e
|
File details
Details for the file pymatching-2.3.1-cp311-cp311-macosx_11_0_arm64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp311-cp311-macosx_11_0_arm64.whl
- Upload date:
- Size: 381.3 kB
- Tags: CPython 3.11, macOS 11.0+ ARM64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
020a7bd1f8a2d312fe98f4eee594cff98395272ab868f93ee202adbdec1901bf
|
|
| MD5 |
97ebd1e2bbe540b2b8c4c50a295902f6
|
|
| BLAKE2b-256 |
9b94eee399cae91c349481467b03189832277ea79781f331cc04bde51834a1e1
|
File details
Details for the file pymatching-2.3.1-cp310-cp310-win_amd64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp310-cp310-win_amd64.whl
- Upload date:
- Size: 345.4 kB
- Tags: CPython 3.10, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b902eab5d530990eb577eabafc8969115252baa48eb9e9c7734b6bca97edc98c
|
|
| MD5 |
196305ece0f7c473ec478915ccbe6823
|
|
| BLAKE2b-256 |
ce226757dc12b3adb5fc97adc414bee2fe63ce66fb534548288ef2d7e6b326f0
|
File details
Details for the file pymatching-2.3.1-cp310-cp310-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp310-cp310-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl
- Upload date:
- Size: 623.8 kB
- Tags: CPython 3.10, manylinux: glibc 2.27+ x86-64, manylinux: glibc 2.28+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
fba6c1320dcad0268073cbba3b5e7296d94c61944a798cf8674cbaef68d9af91
|
|
| MD5 |
2ee71915100d0a4d635da8e1b07ecf80
|
|
| BLAKE2b-256 |
c43cecdc80293a45b4702a8fdb0630ebca9e93efd91a3b6b6986a81be9a5a3a1
|
File details
Details for the file pymatching-2.3.1-cp310-cp310-macosx_11_0_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp310-cp310-macosx_11_0_x86_64.whl
- Upload date:
- Size: 411.6 kB
- Tags: CPython 3.10, macOS 11.0+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
403b4abba5d54ea8012a5d7334a8f6ba871f72875e072753bb7078e81a4f32e4
|
|
| MD5 |
3acbc836a7c3a9a388d01c745f3d743f
|
|
| BLAKE2b-256 |
b44babc76305247b2204eb2a72239c3032647a31d69eb99ca63181027011d7a7
|
File details
Details for the file pymatching-2.3.1-cp310-cp310-macosx_11_0_arm64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp310-cp310-macosx_11_0_arm64.whl
- Upload date:
- Size: 380.0 kB
- Tags: CPython 3.10, macOS 11.0+ ARM64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9f3bd434808ce6dc59730bed2fbb22bb2cc5454f13f4833ba621240e0d75d4f6
|
|
| MD5 |
22ecca1abafd18645ca987a294a7bfee
|
|
| BLAKE2b-256 |
92654e414c1a09b198b5ab771a9249cbbfe47c29951914e1393f787aaf8041b4
|
File details
Details for the file pymatching-2.3.1-cp39-cp39-win_amd64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp39-cp39-win_amd64.whl
- Upload date:
- Size: 348.5 kB
- Tags: CPython 3.9, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
fc41428551840048cc72d2109a837da7d69c016680c1ef9431b12622f4cc00e6
|
|
| MD5 |
9d8890ee3ba7f77e5cc59569f61e2eb1
|
|
| BLAKE2b-256 |
c421d09fd8ae48b87831482adb405f589f5a43de964544452badae00103d75bd
|
File details
Details for the file pymatching-2.3.1-cp39-cp39-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp39-cp39-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl
- Upload date:
- Size: 624.1 kB
- Tags: CPython 3.9, manylinux: glibc 2.27+ x86-64, manylinux: glibc 2.28+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
29314df027ba4915c96d3efb973d49232fea2353f64190d5700a0853a6ed4ef5
|
|
| MD5 |
c500634e0f7c20580c5e1d57ee13b830
|
|
| BLAKE2b-256 |
ea6f36bee02b9c9827b29afbe2e092bfc7507de9fcc195f601ed64d7eb5962c1
|
File details
Details for the file pymatching-2.3.1-cp39-cp39-macosx_11_0_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp39-cp39-macosx_11_0_x86_64.whl
- Upload date:
- Size: 411.8 kB
- Tags: CPython 3.9, macOS 11.0+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2b27c47cd82525fdc7070f20e87c915d415e22d882979ee8ac958ffb6b7571ca
|
|
| MD5 |
5ad7d381b06b60d9387b99eec59c81a9
|
|
| BLAKE2b-256 |
1eb0fe34acc8a4a960978e2ba51502a8c05e4b024befedef2ea7488a41a29f43
|
File details
Details for the file pymatching-2.3.1-cp39-cp39-macosx_11_0_arm64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp39-cp39-macosx_11_0_arm64.whl
- Upload date:
- Size: 380.1 kB
- Tags: CPython 3.9, macOS 11.0+ ARM64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
6073045e8c0768f52a05f62f4976610deb738dc42e75aa2078e77a79a05afb8b
|
|
| MD5 |
faa673e3d3a51f297d7103afb9e59bd0
|
|
| BLAKE2b-256 |
fb1e62c519bce5e8da8f51f1f34b9f99174842c3805d7d379ef4a6234f163fdd
|
File details
Details for the file pymatching-2.3.1-cp38-cp38-win_amd64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp38-cp38-win_amd64.whl
- Upload date:
- Size: 345.2 kB
- Tags: CPython 3.8, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
0c41340f97f909fb5f2ed90b64d3fc96717f60da0122673dcb557c45d7db275e
|
|
| MD5 |
d345d80eafbf4ee23d9bbcf1ae7ac509
|
|
| BLAKE2b-256 |
a02486c72bf8ef2f8b6e2b8dc02edacb657e1b4cb0e9a9f95e28daa5a10d5a46
|
File details
Details for the file pymatching-2.3.1-cp38-cp38-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp38-cp38-manylinux_2_27_x86_64.manylinux_2_28_x86_64.whl
- Upload date:
- Size: 623.6 kB
- Tags: CPython 3.8, manylinux: glibc 2.27+ x86-64, manylinux: glibc 2.28+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
8516b99bda3f5abef9afac5d395f955ddccb511df1a8d18160a72fd771ecce51
|
|
| MD5 |
e6f17566c4cc72dd90d461c2271066ea
|
|
| BLAKE2b-256 |
81b783b14103c761aff6e86b9c7d689fc04ac7808ae4676edb5e3f98937c0f4f
|
File details
Details for the file pymatching-2.3.1-cp38-cp38-macosx_11_0_x86_64.whl.
File metadata
- Download URL: pymatching-2.3.1-cp38-cp38-macosx_11_0_x86_64.whl
- Upload date:
- Size: 411.4 kB
- Tags: CPython 3.8, macOS 11.0+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
dcd0c34adb2966ef48fcb7245e9d929e41b88bbabf3aef4270cb63c36532c6ab
|
|
| MD5 |
7b9337c82249a9143a81ffc2a5e6b3ec
|
|
| BLAKE2b-256 |
d95424e72d71cd9fa184b61f5d2b3830281b4943e88cd018f1c1e6cc6938ff23
|