Simulate DNA band patterns for gel migration experiments
Project description
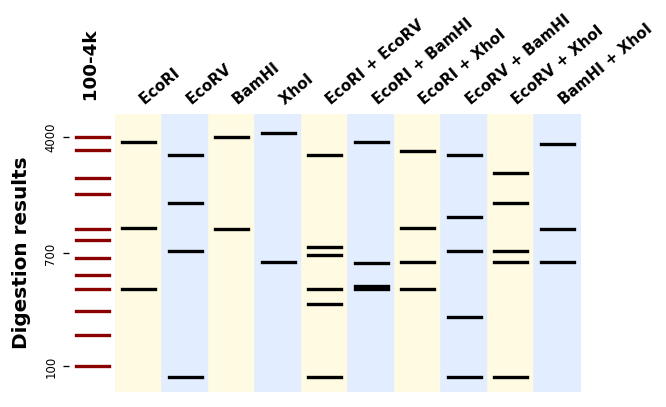
Bandwagon (full documentation here) is a Python library to predict and plot migration patterns from DNA digestions. It supports hundreds of different enzymes (thanks to BioPython), single- and multiple-enzymes digestions, and custom ladders.
It uses Matplotlib to produce plots like this one:
Infos
PIP installation:
pip install bandwagonWeb documentation:
https://edinburgh-genome-foundry.github.io/bandwagon/
Github Page
https://github.com/Edinburgh-Genome-Foundry/bandwagon
Live demo
https://cuba.genomefoundry.org/predict-digestions
License: MIT, Copyright Edinburgh Genome Foundry
More biology software

Bandwagon is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file bandwagon-0.3.1.tar.gz.
File metadata
- Download URL: bandwagon-0.3.1.tar.gz
- Upload date:
- Size: 21.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.5.0.1 requests/2.24.0 setuptools/50.1.0 requests-toolbelt/0.9.1 tqdm/4.48.0 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5e4f8319dedc58daa9012c34bcaed035f306f07aedc507bfcb05e3f8e2d5aea0
|
|
| MD5 |
6ae957e1eff1468a0ef11dbd755311cf
|
|
| BLAKE2b-256 |
389bed4eb1f9686ddb17e2ff061c5395b5d6b13a80184eed39df9d1ad690132b
|
File details
Details for the file bandwagon-0.3.1-py3-none-any.whl.
File metadata
- Download URL: bandwagon-0.3.1-py3-none-any.whl
- Upload date:
- Size: 17.3 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.5.0.1 requests/2.24.0 setuptools/50.1.0 requests-toolbelt/0.9.1 tqdm/4.48.0 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
cea52890e70e54695a4816a6414d001ba59c98916a64896282a1eb84cd93b6d5
|
|
| MD5 |
f7d835d72d5341569346f69935d98a90
|
|
| BLAKE2b-256 |
4bed3e379de7a4cf2864bbaff3e4f7dc78f97298a68e8d62fdcf682932a09a4a
|











