Efficient cell detection in large images
Project description
cellfinder-napari
Efficient cell detection in large images (e.g. whole mouse brain images)
This package implements the cell detection algorithm from Tyson, Rousseau & Niedworok et al. (2021) for napari, based on the cellfinder-core package.
This algorithm can also be used within the original cellfinder software for whole-brain microscopy analysis.
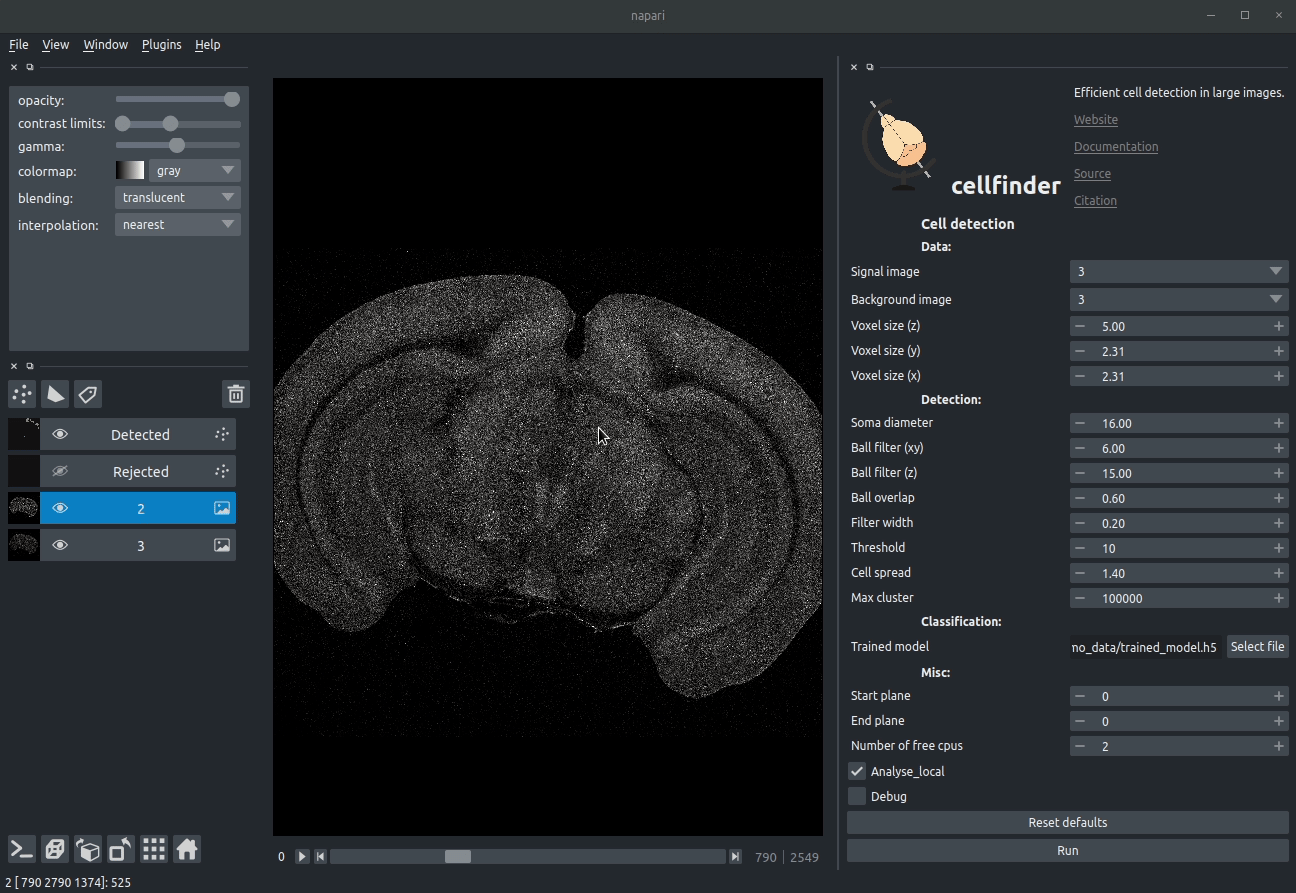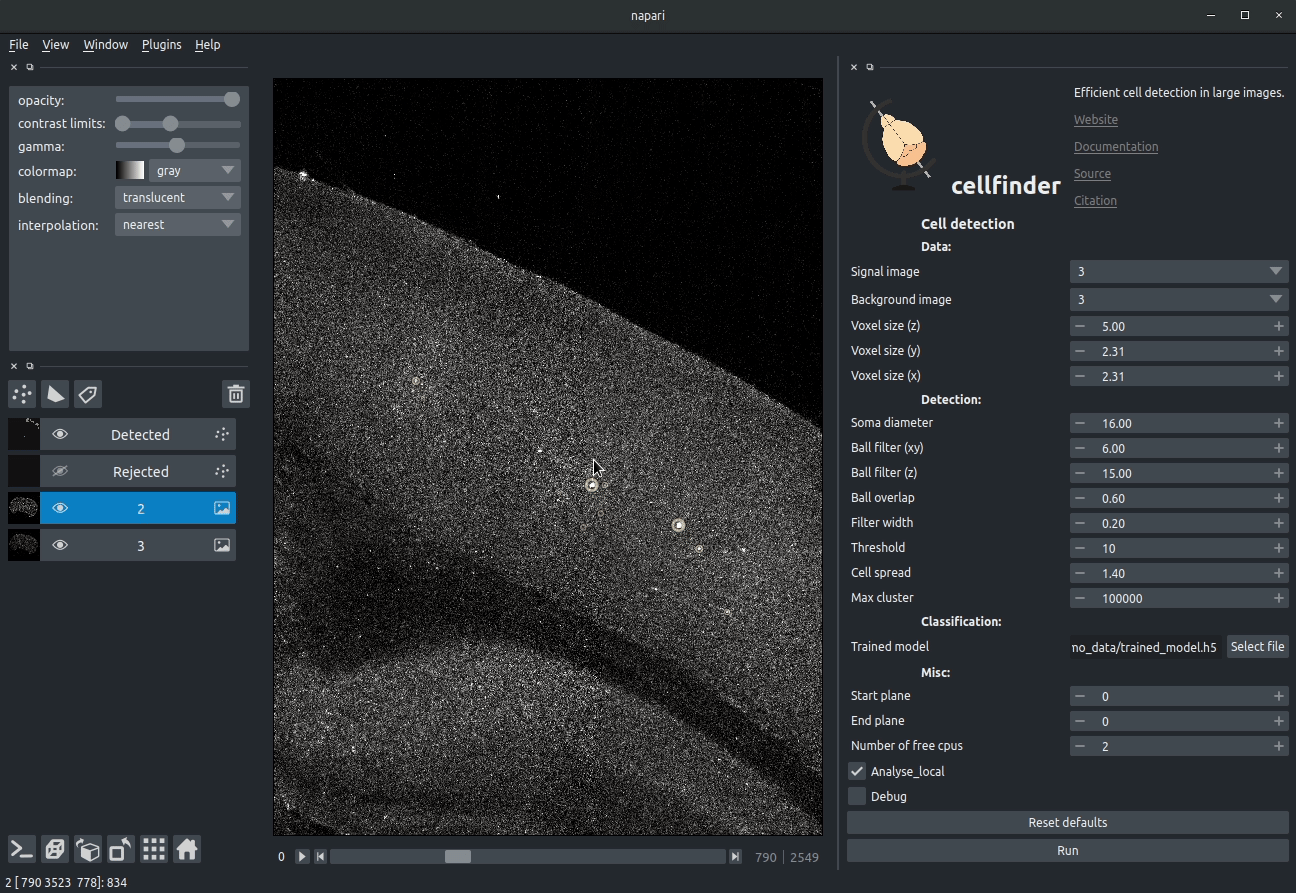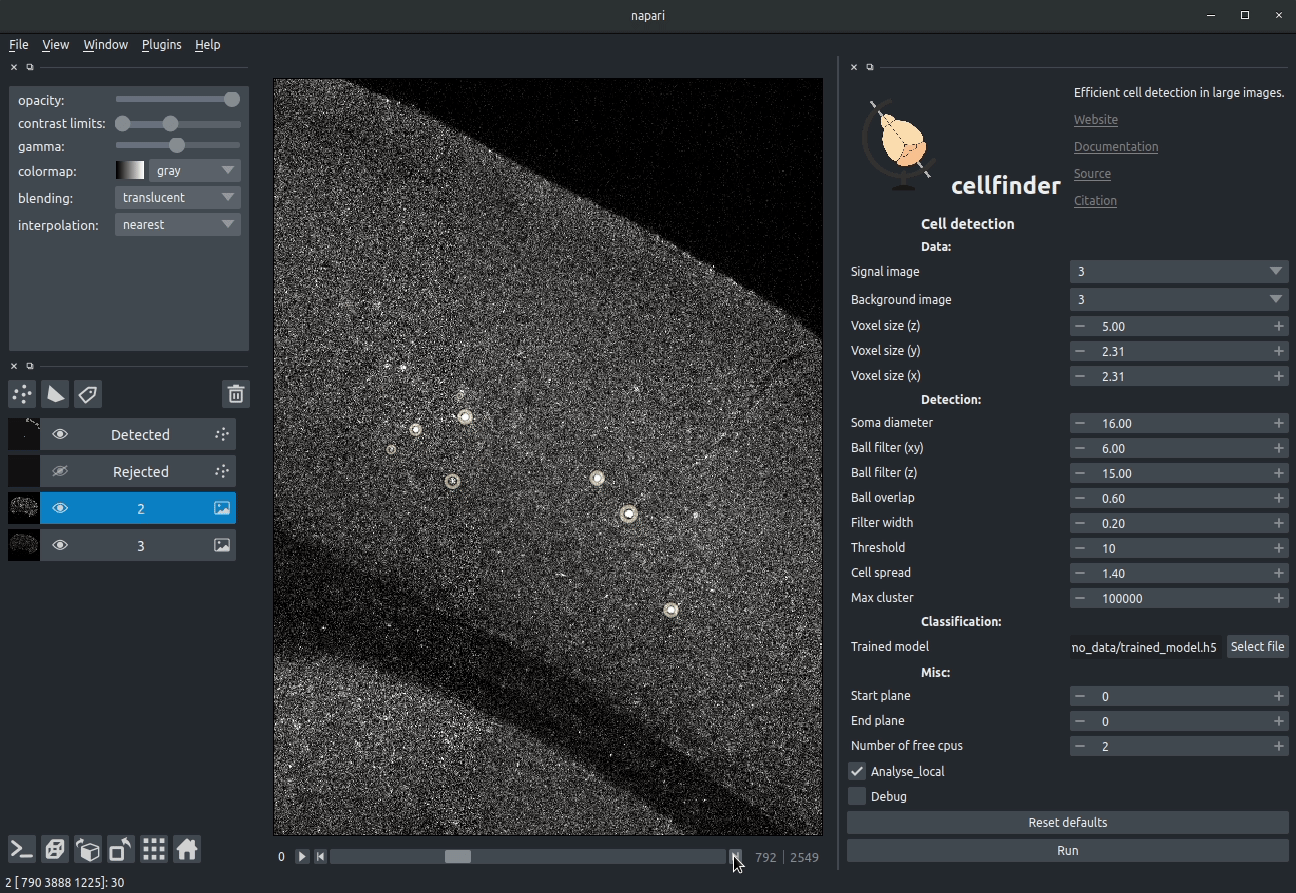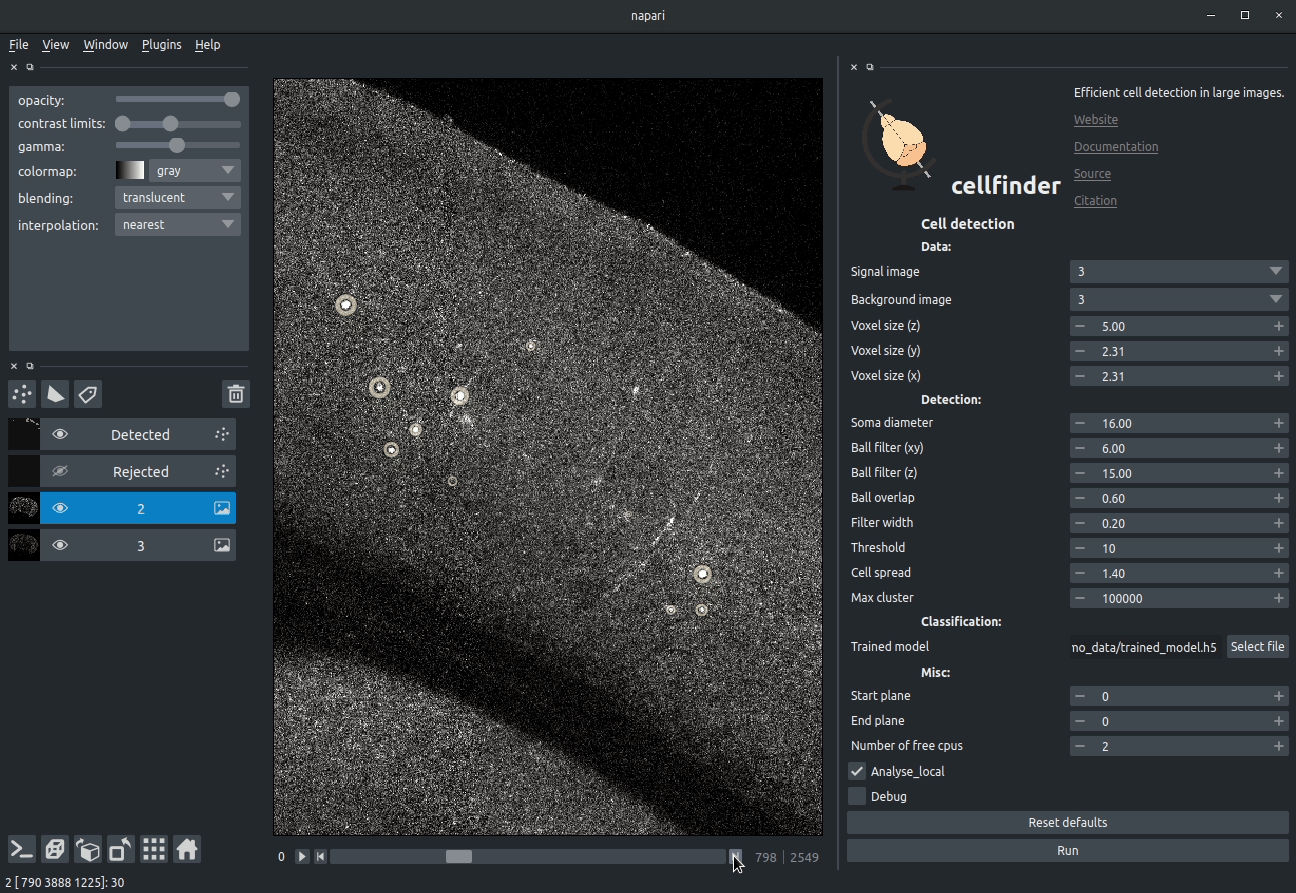
Visualising detected cells in the cellfinder napari plugin
Instructions
Installation
Once you have installed napari. You can install napari either through the napari plugin installation tool, or directly from PyPI with:
pip install cellfinder-napari
Usage
Full documentation can be found here.
This software is at a very early stage, and was written with our data in mind. Over time we hope to support other data types/formats. If you have any questions or issues, please get in touch by email, gitter or by raising an issue.
Illustration
Introduction
cellfinder takes a stitched, but otherwise raw whole-brain dataset with at least two channels:
- Background channel (i.e. autofluorescence)
- Signal channel, the one with the cells to be detected:

Cell candidate detection
Classical image analysis (e.g. filters, thresholding) is used to find cell-like objects (with false positives):

Cell candidate classification
A deep-learning network (ResNet) is used to classify cell candidates as true cells or artefacts:

Citing cellfinder
If you find this plugin useful, and use it in your research, please cite the preprint outlining the cell detection algorithm:
Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ bioRxiv, doi.org/10.1101/2020.10.21.348771
If you use this, or any other tools in the brainglobe suite, please let us know, and we'd be happy to promote your paper/talk etc.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file cellfinder-napari-0.0.5.tar.gz.
File metadata
- Download URL: cellfinder-napari-0.0.5.tar.gz
- Upload date:
- Size: 15.8 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/4.0.1 pkginfo/1.7.0 requests/2.25.1 requests-toolbelt/0.9.1 tqdm/4.60.0 CPython/3.9.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c79d441e5e84371c93928df8103d17a148ff440896a7c3a5fcb000b4fd9d3c56 |
|
| MD5 | 9abe2a007b30c656608b5e14ab8f1257 |
|
| BLAKE2b-256 | 2e969355fea9653b4ab24d6fd353439b15fca9fe097934d0a1cd8e7ecf86dcfe |
File details
Details for the file cellfinder_napari-0.0.5-py3-none-any.whl.
File metadata
- Download URL: cellfinder_napari-0.0.5-py3-none-any.whl
- Upload date:
- Size: 15.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/4.0.1 pkginfo/1.7.0 requests/2.25.1 requests-toolbelt/0.9.1 tqdm/4.60.0 CPython/3.9.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b90dd9a202becfbd232213706d4b344a722a9889fb31db4ca2370dba8f2ae3a2 |
|
| MD5 | 7d8e1660253fc42651c8c316e38cc49d |
|
| BLAKE2b-256 | 090b28fa2f1111fda5b5ecbf9b7b600042400a791812e0c9ac1ad1f0863f7a44 |