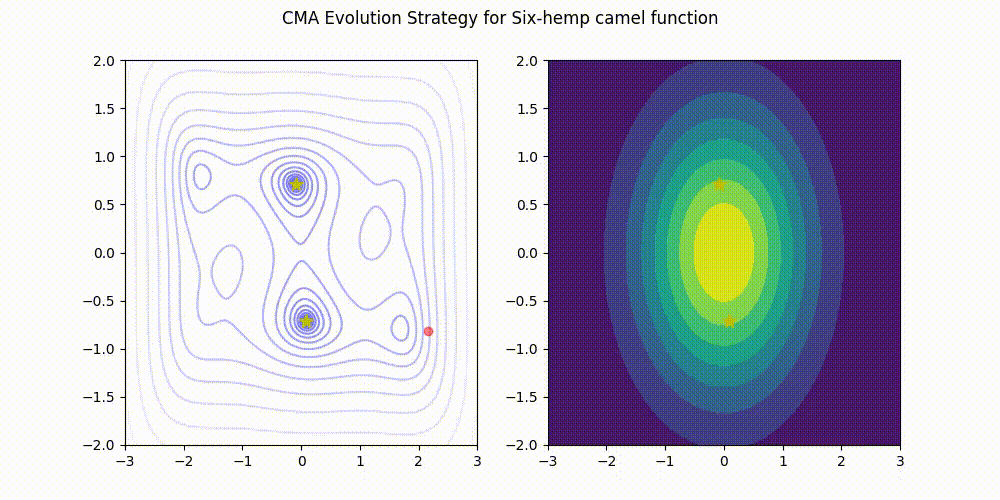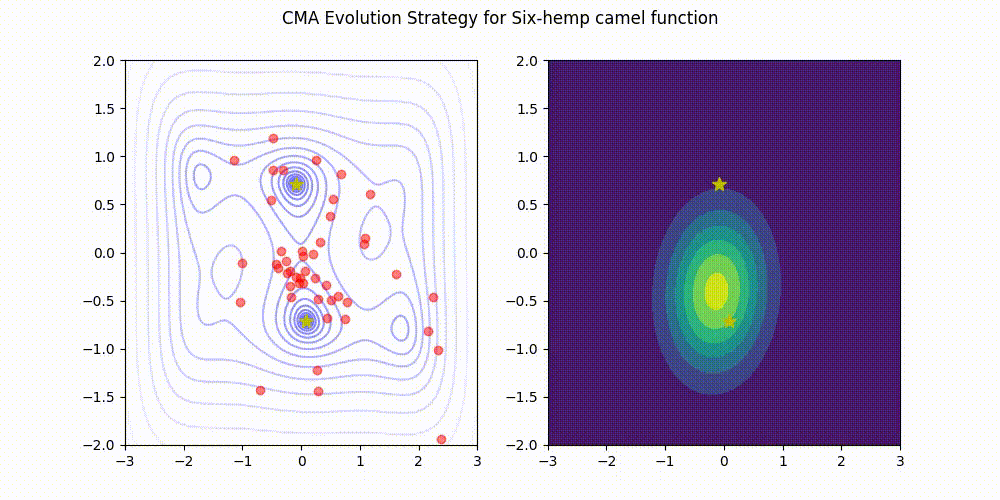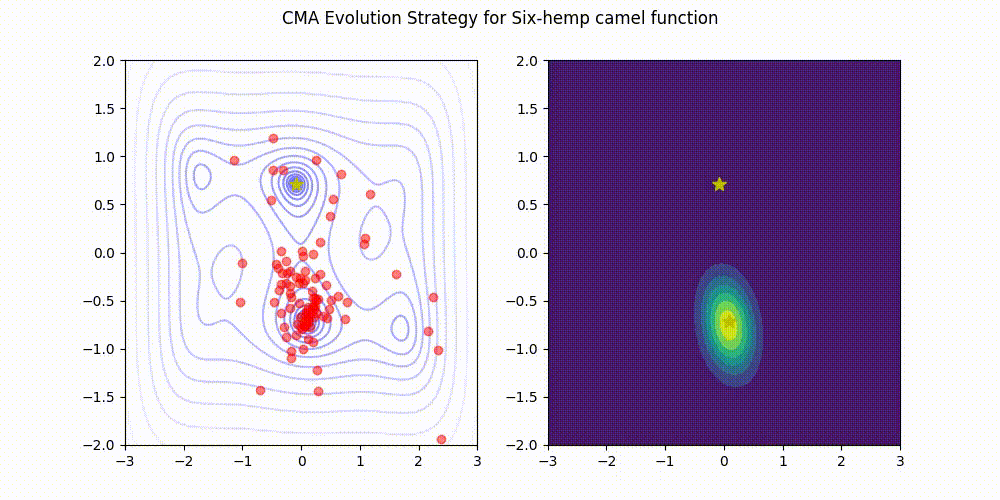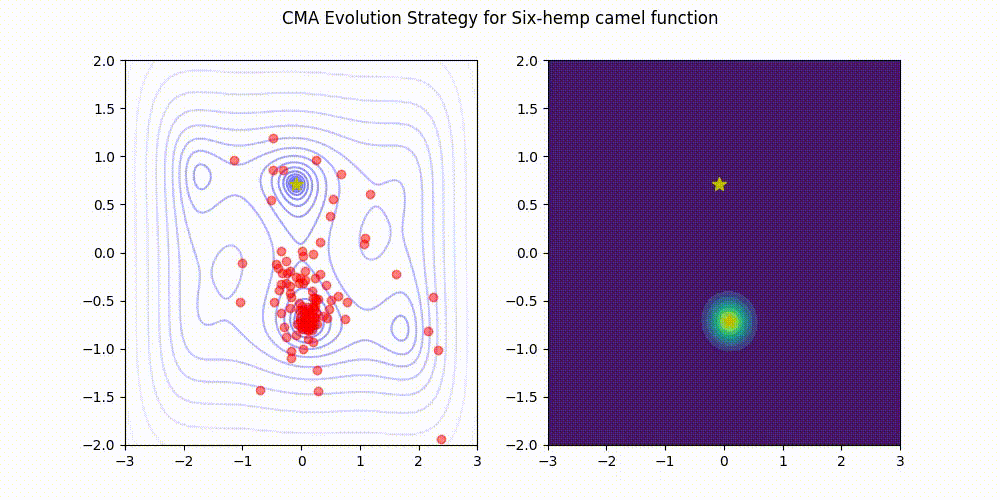
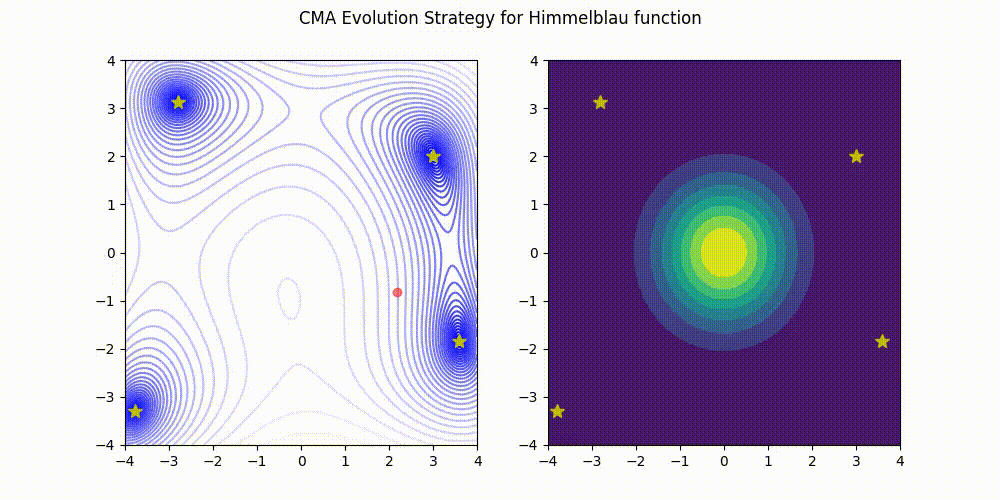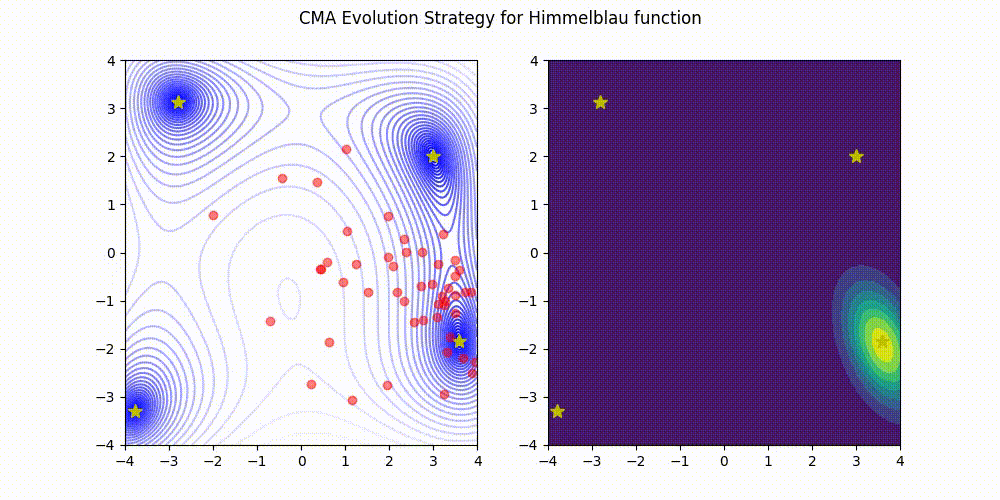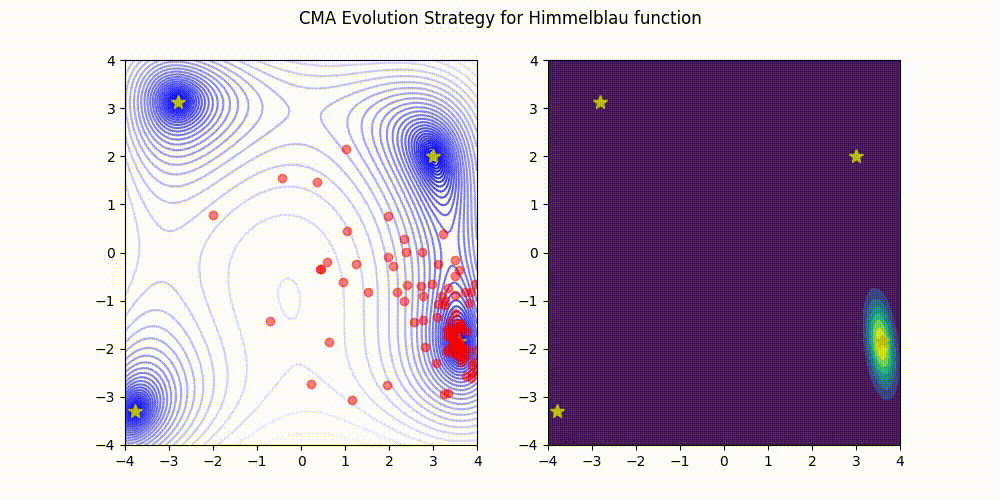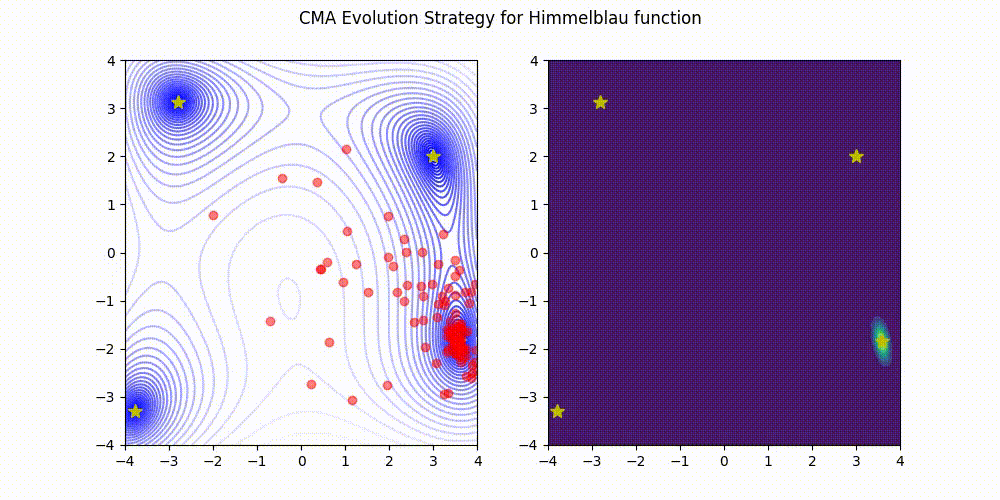
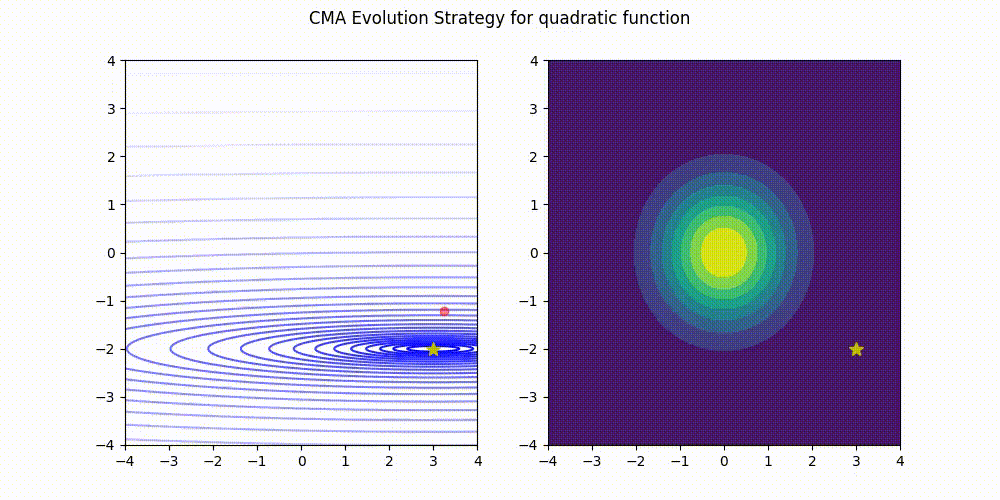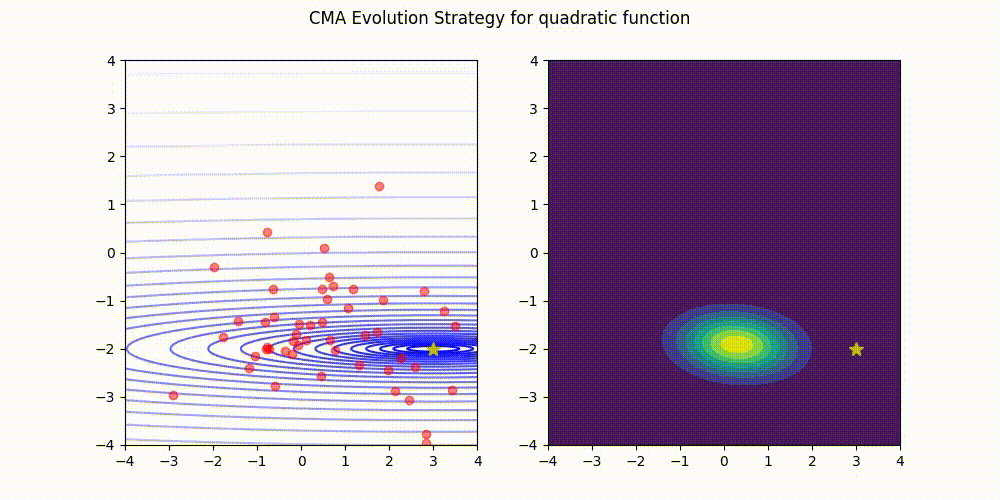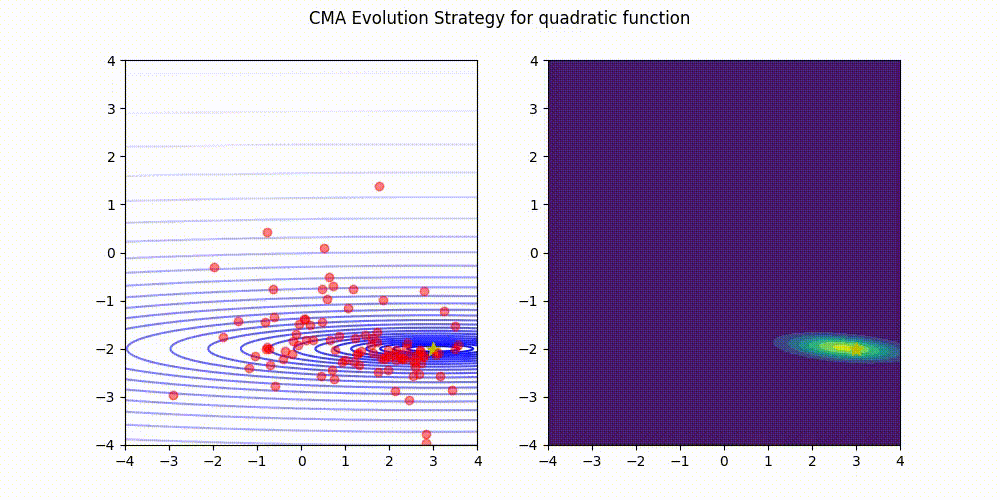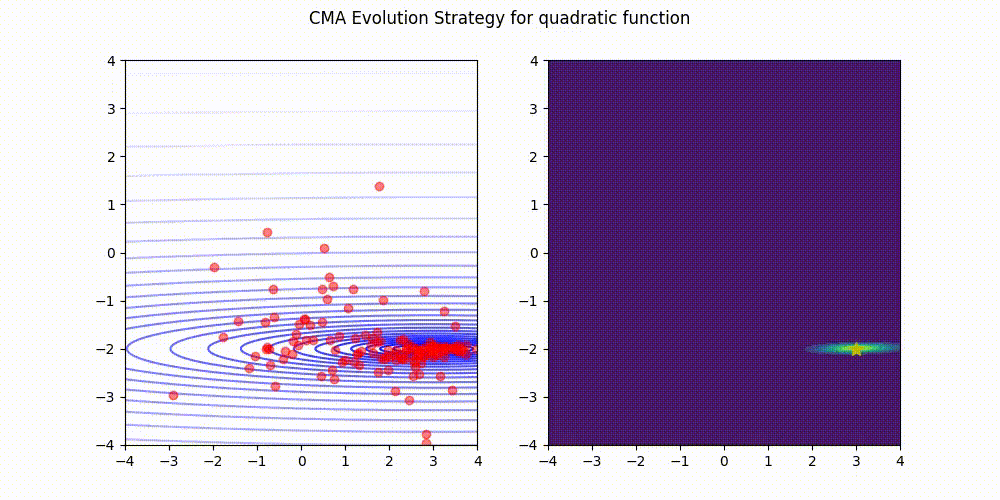
Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) implementation for Python 3.
Project description
CMA-ES
Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) [1] implementation.
Himmelblau function.
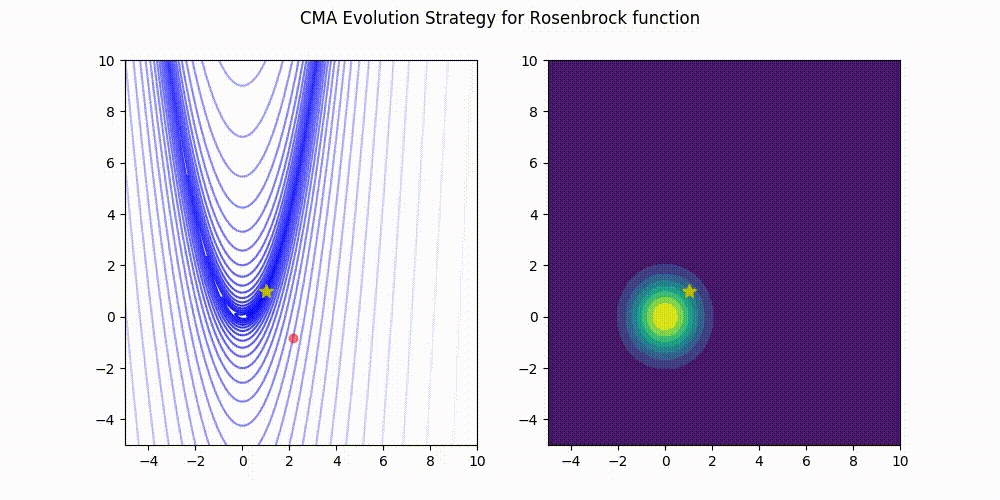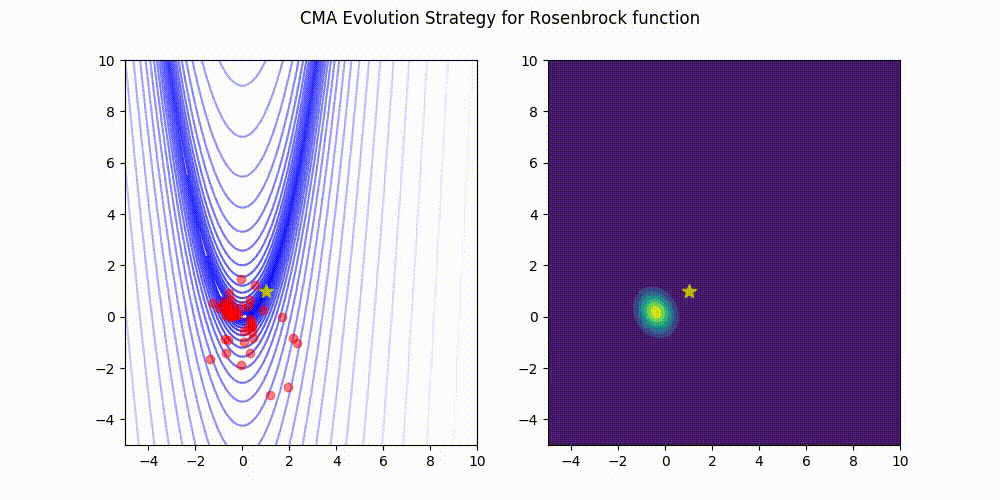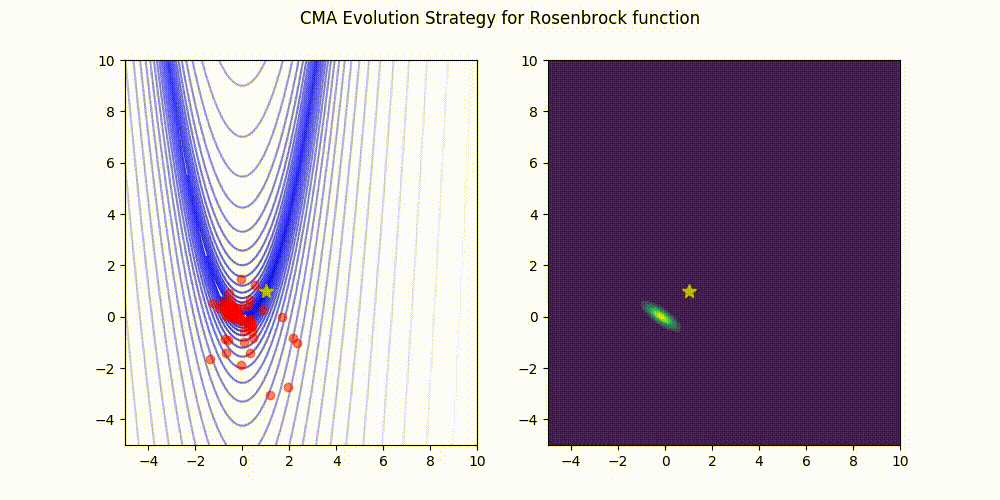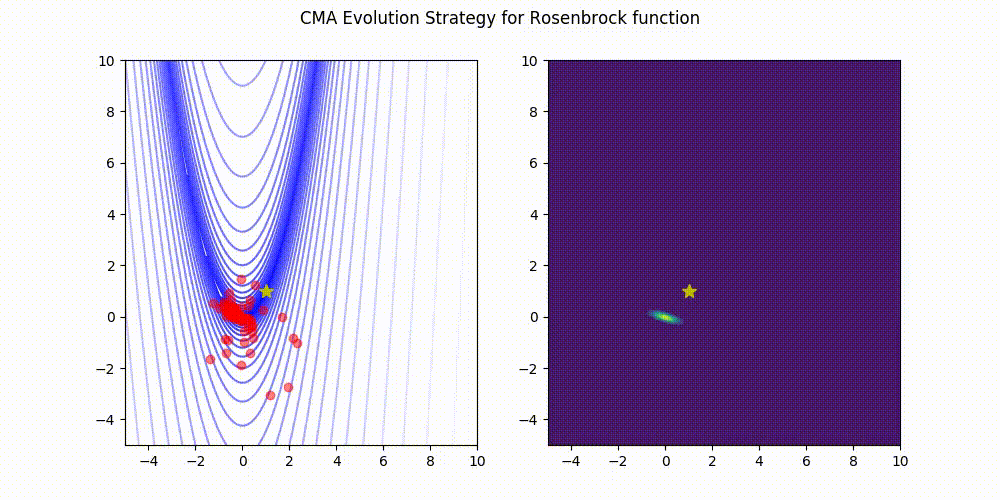
Rosenbrock function.
Quadratic function.
These GIF animations are generated by visualizer.py.
Installation
Supported Python versions are 3.6 or later.
$ pip install cmaes
Or you can install via conda-forge.
$ conda install -c conda-forge cmaes
Usage
This library provides two interfaces that an Optuna's sampler interface and a low-level interface. I recommend you to use this library via Optuna.
Optuna's sampler interface
Optuna [2] is an automatic hyperparameter optimization framework. A sampler based on this library is available from Optuna v1.3.0. Usage is like this:
import optuna
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = optuna.samplers.CmaEsSampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)
See the documentation for more details.
Monkeypatch for faster CMA-ES sampler of Optuna v1.3.x.
If you are using Optuna v1.3.x, you can make optuna.samplers.CmaEsSampler faster.
import optuna
from cmaes.monkeypatch import patch_fast_intersection_search_space
patch_fast_intersection_search_space()
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = optuna.samplers.CmaEsSampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)
For older versions (Optuna v1.2.0 or older)
If you are using older versions, please use cmaes.samlper.CMASampler.
import optuna
from cmaes.sampler import CMASampler
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = CMASampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)
Note that CMASampler doesn't support categorical distributions. Although pycma's sampler supports categorical distributions, it also has a problem (especially on high-cardinality categorical distribution). If your search space contains a categorical distribution, please use TPESampler.
Low-level interface
This library also provides an "ask-and-tell" style interface.
import numpy as np
from cmaes import CMA
def quadratic(x1, x2):
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
cma_es = CMA(mean=np.zeros(2), sigma=1.3)
for generation in range(50):
solutions = []
for _ in range(cma_es.population_size):
x = cma_es.ask()
value = quadratic(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
cma_es.tell(solutions)
Benchmark results
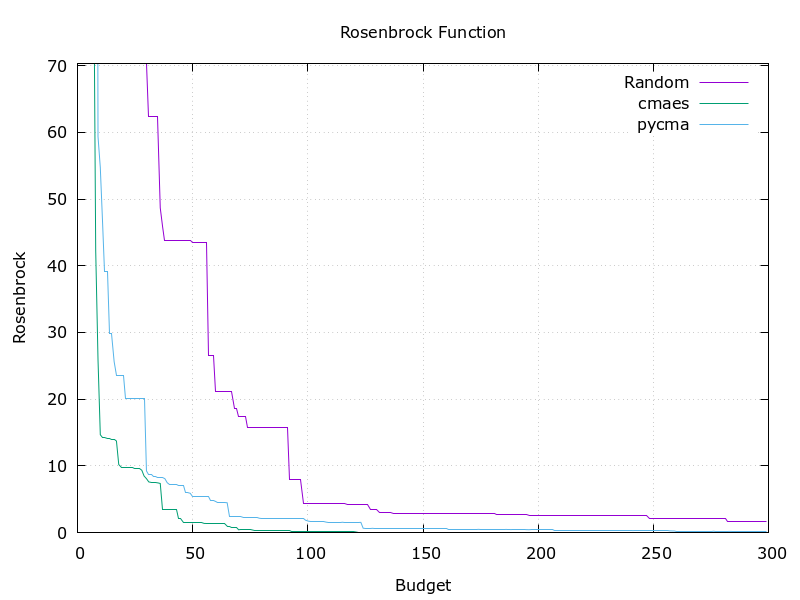
Optuna officially implements a sampler based on pycma. It achieves almost the same performance. But this library is faster and simple.
Algorithm's efficiency
| Rosenbrock function | Six-Hemp Camel function |
|---|---|
 |
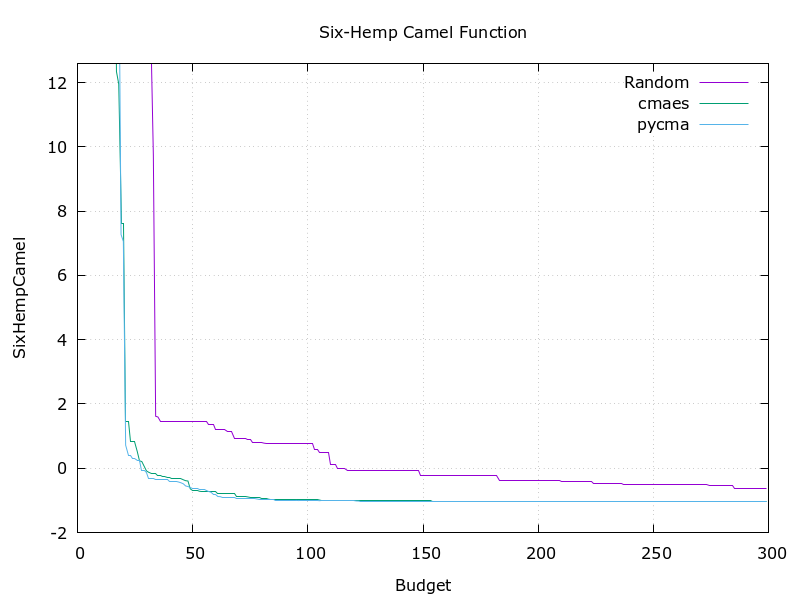 |
This implementation (green) stands comparison with pycma (blue). See benchmark for details.
Execution Speed
| trials/params | storage | pycma integration sampler | this library |
|---|---|---|---|
| 100 / 5 | memory | 4.976 sec (+/- 0.596) | 0.197 sec (+/- 0.078) |
| 500 / 5 | memory | 71.651 sec (+/- 3.847) | 0.656 sec (+/- 0.044) |
| 500 / 50 | memory | 291.002 sec (+/- 5.010) | 1.981 sec (+/- 0.041) |
| 100 / 5 | sqlite | 16.143 sec (+/- 3.487) | 11.843 sec (+/- 1.390) |
| 500 / 5 | sqlite | 129.436 sec (+/- 6.279) | 43.735 sec (+/- 2.676) |
| 500 / 50 | sqlite | 397.084 sec (+/- 6.618) | 150.531 sec (+/- 1.113) |
This script was run on my laptop with --times 4. So the times should not be taken precisely.
Even though, it is clear that this library is extremely faster than Optuna's pycma sampler (with Optuna v1.0.0 and pycma v2.7.0).
Links
Other libraries:
I respect all libraries involved in CMA-ES.
- pycma : Most famous CMA-ES implementation by Nikolaus Hansen.
- cma-es : A Tensorflow v2 implementation.
References:
- [1] N. Hansen, The CMA Evolution Strategy: A Tutorial. arXiv:1604.00772, 2016.
- [2] Takuya Akiba, Shotaro Sano, Toshihiko Yanase, Takeru Ohta, Masanori Koyama. 2019. Optuna: A Next-generation Hyperparameter Optimization Framework. In The 25th ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD ’19), August 4–8, 2019.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file cmaes-0.4.0.tar.gz.
File metadata
- Download URL: cmaes-0.4.0.tar.gz
- Upload date:
- Size: 15.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.23.0 setuptools/41.2.0 requests-toolbelt/0.9.1 tqdm/4.45.0 CPython/3.8.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
791c5fb0b2ec1a3bcd873120bcadfadd60dca1c2f75adcba1b2007f977f707cc
|
|
| MD5 |
ed7b3c31bfdbd1575a79f16dad1cc332
|
|
| BLAKE2b-256 |
72671a627614d62ea8bc3e4d84c558a4be861956db4007b61b98c0bb5bf7ec39
|
File details
Details for the file cmaes-0.4.0-py3-none-any.whl.
File metadata
- Download URL: cmaes-0.4.0-py3-none-any.whl
- Upload date:
- Size: 13.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.23.0 setuptools/41.2.0 requests-toolbelt/0.9.1 tqdm/4.45.0 CPython/3.8.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
ce8914ba36fe174621ed05c4fd837301e20c742d49314dfbf42e6654f365c096
|
|
| MD5 |
b1abd1b0c6efe70cb718c571a4c6f7c2
|
|
| BLAKE2b-256 |
560016b9a086181cd227178d5279a40f7dce4fd16d7b3f6bdbd986b38d6bd3f0
|