Skeletonize densely labeled image volumes.
Project description
fill_voids - High performance binary_fill_holes
import fill_voids
img = ... # 3d binary image
filled_image = fill_voids.fill(img, in_place=False) # in_place allows editing of original image
3D void filling algorithm, similar function as scipy.ndimage.morphology.binary_fill_holes
The purpose of this repo is to make it convenient to improve upon the scipy hole filling algorithm which only applies to binary images and uses slow serial dilations.
The current version of the improvements is to use a scan line flood fill of the background and label everything not filled as foreground. This is already significantly faster and lower memory, but we can do better by reducing memory usage and supporting multiple labels.
Current version
- Allocate a uint8 zeroed 3D image that is 2 voxels larger than the binary input image along each dimension.
- Paint the input image into the new image such that there is a black border around it setting foreground as 2.
- Flood fill (six connected) from a spot on the border (arbitarily taken to be (0,0,0)) and label the flood fill as 1.
- Write out a binary image the same size as the input from the temporary buffer where foreground is set as buffer != 1 (i.e. 0 or 2).
We improve performance significantly by using libdivide to make computing x,y,z coordinates from array index faster, by scanning right and left to take advantage of machine memory speed, by only placing a neighbor on the stack when we've either just started a scan or just passed a foreground pixel while scanning.
Binary Version Improvements
It would be possible to skip the allocating and painting steps by walking along each side of the image and adding newly encountered voids as seed points to the stack. This reduces the memory usage to near zero.
Multi-Label Improvements
Similarly to the connected-components-3d and euclidean-distance-3d projects, in connectomics, it can be common to want to apply void filling algorithms to all labels within a densely packed volume. A multi-label algorithm can be much faster than even the fastest serial application of a binary algorithm. Here's how this might go given an input image I:
- Compute M = max(I)
- Perform the fill as in the binary algorithm labeling the surrounding void as M+1. This means all voids are now either legitimate and can be filled or holes in-between labels.
- Raster scan through the volume. If a new void is encountered, we must determine if it is fillable or an in-between one which will not be filled.
- On encountering the void, record the last label seen and contour trace around it. If only that label is encountered during contour tracing, it is fillable. If another label is encountered, it is not fillable.
- During the contour trace, mark the trace using an integer not already used, such as M+2. If that label is encountered in the future, you'll know what to fill between it and the next label encountered based on the fillable determination. This phase stops when either the twin of the first M+2 label is encountered or when futher contour tracing isn't possible (in the case of single voxel gaps).
- (Inner Labels) If another label is encountered in the middle of a void, contour trace around it and mark the boundary with the same M+2 label that started the current fill.
SciPy Comparison
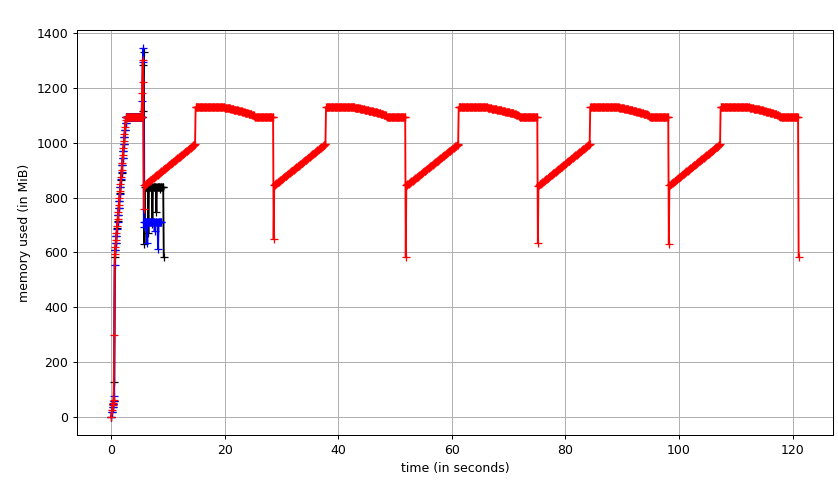
Fig. 1: Filling five labels using SciPy binary_fill_holes vs fill_voids from a 512x512x512 densely labeled connectomics segmentation. (black) fill_voids 0.1 (blue) scipy 1.3.3
In this test, fill_voids is significantly faster than scipy at lower memory with in_place=False.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distributions
Hashes for fill_voids-1.0.0-cp38-cp38-win_amd64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 94deb7c5069fd739d96f19d891de2feb5587cf9d0e25ffec6cdcdaac4a0e8892 |
|
| MD5 | bf89d8b08cc935fd5b3c2b24352fa413 |
|
| BLAKE2b-256 | 71bbc39a84303f818faacd7b7d85fad52770ad920764161f86d85b7132bce943 |
Hashes for fill_voids-1.0.0-cp38-cp38-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 48342fa2c1bb15b59da41c0aaed66311ae6c3a8fb9c9dbe0a3f5de9d192ec569 |
|
| MD5 | 79f01922510644df7af52237eea5c7d4 |
|
| BLAKE2b-256 | 15a7b91382443d430155484918cc28e918a5b18d403f3c5c26aa1a62ed65ed04 |
Hashes for fill_voids-1.0.0-cp38-cp38-macosx_10_9_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ffe456425746b82e6a03b38918c3725fd4cec6c6aca2a6ef72678b4a2501632f |
|
| MD5 | be2f421a18e73f79fff0cb5ea758ab34 |
|
| BLAKE2b-256 | f88b10dfa34b2821f0d18c9de55ab4cf7bb8093288715570e50dc6872650fef5 |
Hashes for fill_voids-1.0.0-cp37-cp37m-win_amd64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c458ffb8ef28d515624ccc7ab5ed870c9a0a2b6b1eaaaf54e673934f91856a1b |
|
| MD5 | 2016b8d14421f8951456d400bdb86356 |
|
| BLAKE2b-256 | 3cb9678e2d7aad3785e53e00fe45c14f96f96a60d2296992905ce04c0eeb7c45 |
Hashes for fill_voids-1.0.0-cp37-cp37m-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 5099e82d57be46811a330885cacc90361ee17daf7bd16479646455a38479f0af |
|
| MD5 | 19c4580ee1f3218299cd688555d21403 |
|
| BLAKE2b-256 | d11b56c170bf23aecc6e96a9c90afc67ac6747fa2db6c695263f7c560c7138de |
Hashes for fill_voids-1.0.0-cp37-cp37m-macosx_10_9_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 027d8db52e081c2809a71208cdf794a68bfcadd8a6cb1e23e118fe2a4e074aae |
|
| MD5 | a039c23d1284fbc6645402f30c5a3999 |
|
| BLAKE2b-256 | 9984e9519688f076c937574b200b1ce45dcbbcfcf58d80e0e979fc5a23d5f2a4 |
Hashes for fill_voids-1.0.0-cp36-cp36m-win_amd64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 5396cd031fafd8bf51a219e681e30867ab71877b331e0ca11fa1fd5f87636972 |
|
| MD5 | 2bbb40a8428c9d828ab49085ad209e23 |
|
| BLAKE2b-256 | 9b9a369f5c85c59dc60ffe1bc11c61bdbdc3800eeaeb8876e67221bd6b65063b |
Hashes for fill_voids-1.0.0-cp36-cp36m-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 45e2c915bfe28c56e86c4cd9a896cec0997eb57770a4c7b550e683ae8df79eee |
|
| MD5 | 5e807aa5ec5a6e90d86a98c916aa6d44 |
|
| BLAKE2b-256 | 9bf7d707d72783cee5b80bdee878cb46f7986f8e26879903b6654556e66fc2cf |
Hashes for fill_voids-1.0.0-cp36-cp36m-macosx_10_9_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 66440f8d79420871dfb284d2e997ffcf1f841141c1251c40b175af516ac2a679 |
|
| MD5 | 42b8ddfb138de34ca3388e0e1b007bb6 |
|
| BLAKE2b-256 | a3e3231723f8cb9991e8d276d0890c112ee3aaf7c92f785fa566134d5638885f |
Hashes for fill_voids-1.0.0-cp35-cp35m-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 2c164fb72c4c9d1af8b75a962951b5bd6159e5be72a2ee1e8cc7e8df6110553f |
|
| MD5 | 44351e6d0c0678e5dea16acc75b3b6aa |
|
| BLAKE2b-256 | dda86fc83c1b04d0fc6c10193fd0a212867251b0f1dc2f4a7e003b89fadb757e |
Hashes for fill_voids-1.0.0-cp27-cp27m-manylinux1_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 98378e10c3e9ec3bf0a752b5b610a1910e03c89cf44e04ab093d38490b750bf5 |
|
| MD5 | 7897ed842202005f402bb98f568de923 |
|
| BLAKE2b-256 | 4a5397e1e2bbff455bc8205fcd9ad436f0e6e4cc192e03faefae9c766ea0e548 |
Hashes for fill_voids-1.0.0-cp27-cp27m-macosx_10_14_intel.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | a385c6bea4f35d4b5dde67caec71e4f0bc3bffcc74524955ff604b4abefccdcc |
|
| MD5 | 8fc0fd2770b1dfd3ab42b73272b5adee |
|
| BLAKE2b-256 | 426fb27739619cf658dba05a5785cc7cb5abb332b0a6e7c293e2634c46821eed |