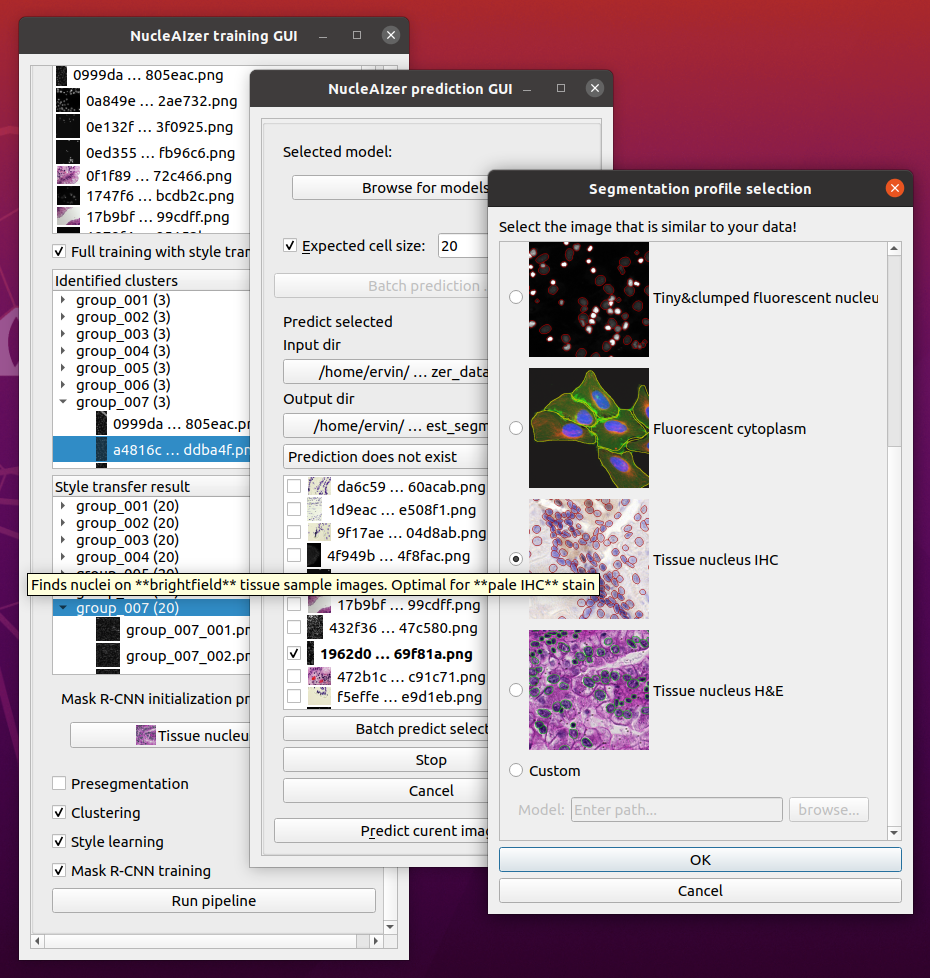
A GUI interface for training and prediction using the nucleAIzer nuclei detection method.
Project description
napari_nucleaizer
GUI for the nucleaAIzer method in Napari.
Overview
This is a napari plugin to execute the nucleaizer nuclei segmentation algorithm.
Main functionalities
Using this plugin will be able to
- Load your image into Napar, then outline the nuclei.
- Specify an image folder containing lots of images and an output folder, and automatically segment all of the images in the input folder.
- If you are not satisfied with the results, you can train your own model:
- You can use our pretrained models and fine tune them on your data.
- You can skip the nucleaizer pipeline and train only on your data.
Supported image types
We have several pretrained models for the following image modelities:
- fluorescent microscopy images
- IHC stained images
- brightfield microscopy images,
among others. For the detailed descriptions of our models, see: https://zenodo.org/record/6790845.
How it works?
For the description of the algorithm, see our paper: "Hollandi et al.: nucleAIzer: A Parameter-free Deep Learning Framework for Nucleus Segmentation Using Image Style Transfer, Cell Systems, 2020. https://doi.org/10.1016/j.cels.2020.04.003"
The original code (https://github.com/spreka/biomagdsb) is partially transformed into a python package (nucleaizer_backend) to actually perform the operations. See the project page of the backend at: https://github.com/etasnadi/nucleaizer_backend.
If you wish to use the web interface, check: http://nucleaizer.org.
Install
-
Create an environment (recommended).
-
Install napari:
pip install "napari[pyqt5]". Other methods: https://napari.org/tutorials/fundamentals/installation.html -
Clone this project and use
pythhon3 -m pip install -e <path>to install the project locally into the same evnrionment as napari.
Run
- Start napari by calling
napari. - Then, activate the plugin in the
Pluginsmenu.
Further help
See the documentation
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
Hashes for napari_nucleaizer-0.2.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 8416b956f46b135e5e19f551ca665ac3e58d4719ec6d7386935b4007d904478d |
|
| MD5 | 75aaef9bd27603ab930414d1ad930ef1 |
|
| BLAKE2b-256 | b4955510dcca023119e5a4a46eba1395e06053a78fd1fcd06f8ee4c11fd6038f |