Multi-class confusion matrix library in Python
Project description
Overview
PyCM is a multi-class confusion matrix library written in Python that supports both input data vectors and direct matrix, and a proper tool for post-classification model evaluation that supports most classes and overall statistics parameters. PyCM is the swiss-army knife of confusion matrices, targeted mainly at data scientists that need a broad array of metrics for predictive models and accurate evaluation of a large variety of classifiers.
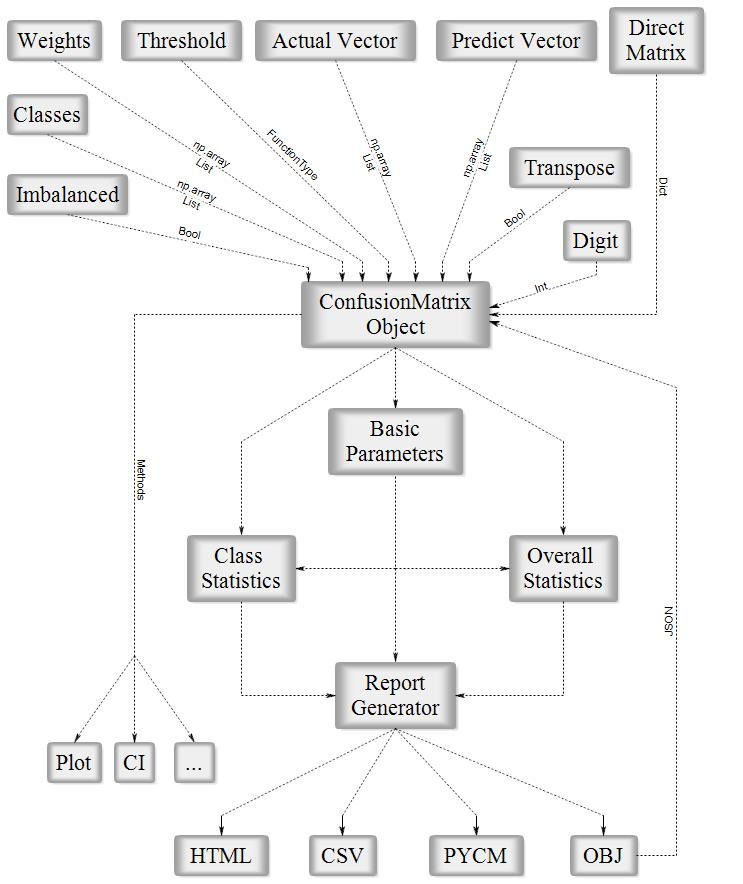
Fig1. ConfusionMatrix Block Diagram
| Open Hub |  |
| PyPI Counter |  |
| Github Stars |  |
| Branch | master | dev |
| CI |  |
 |
| Code Quality |  |
 |
Installation
⚠️ PyCM 4.3 is the last version to support Python 3.6
⚠️ PyCM 3.9 is the last version to support Python 3.5
⚠️ PyCM 2.4 is the last version to support Python 2.7 & Python 3.4
⚠️ Plotting capability requires Matplotlib (>= 3.0.0) or Seaborn (>= 0.9.1)
PyPI
- Check Python Packaging User Guide
- Run
pip install pycm==4.5
Source code
- Download Version 4.5 or Latest Source
- Run
pip install .
Conda
- Check Conda Managing Package
- Update Conda using
conda update conda - Run
conda install -c sepandhaghighi pycm
MATLAB
- Download and install MATLAB (>=8.5, 64/32 bit)
- Download and install Python3.x (>=3.7, 64/32 bit)
- Select
Add to PATHoption - Select
Install pipoption
- Select
- Run
pip install pycm - Configure Python interpreter
>> pyversion PYTHON_EXECUTABLE_FULL_PATH
- Visit MATLAB Examples
Usage
From vector
>>> from pycm import *
>>> y_actu = [2, 0, 2, 2, 0, 1, 1, 2, 2, 0, 1, 2]
>>> y_pred = [0, 0, 2, 1, 0, 2, 1, 0, 2, 0, 2, 2]
>>> cm = ConfusionMatrix(actual_vector=y_actu, predict_vector=y_pred)
>>> cm.classes
[0, 1, 2]
>>> cm.table
{0: {0: 3, 1: 0, 2: 0}, 1: {0: 0, 1: 1, 2: 2}, 2: {0: 2, 1: 1, 2: 3}}
>>> cm.print_matrix()
Predict 0 1 2
Actual
0 3 0 0
1 0 1 2
2 2 1 3
>>> cm.print_normalized_matrix()
Predict 0 1 2
Actual
0 1.0 0.0 0.0
1 0.0 0.33333 0.66667
2 0.33333 0.16667 0.5
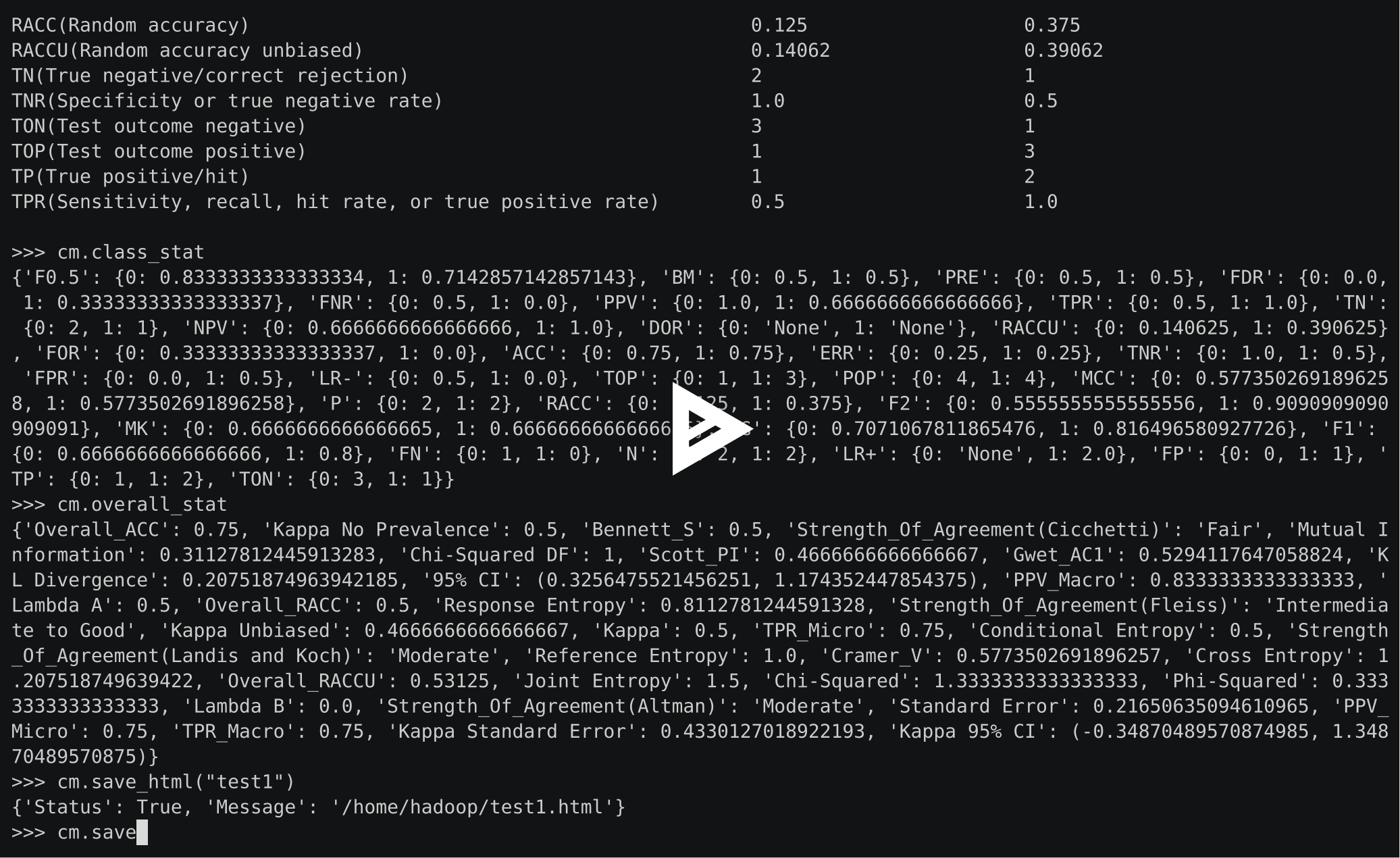
>>> cm.stat(summary=True)
Overall Statistics :
ACC Macro 0.72222
F1 Macro 0.56515
FPR Macro 0.22222
Kappa 0.35484
Overall ACC 0.58333
PPV Macro 0.56667
SOA1(Landis & Koch) Fair
TPR Macro 0.61111
Zero-one Loss 5
Class Statistics :
Classes 0 1 2
ACC(Accuracy) 0.83333 0.75 0.58333
AUC(Area under the ROC curve) 0.88889 0.61111 0.58333
AUCI(AUC value interpretation) Very Good Fair Poor
F1(F1 score - harmonic mean of precision and sensitivity) 0.75 0.4 0.54545
FN(False negative/miss/type 2 error) 0 2 3
FP(False positive/type 1 error/false alarm) 2 1 2
FPR(Fall-out or false positive rate) 0.22222 0.11111 0.33333
N(Condition negative) 9 9 6
P(Condition positive or support) 3 3 6
POP(Population) 12 12 12
PPV(Precision or positive predictive value) 0.6 0.5 0.6
TN(True negative/correct rejection) 7 8 4
TON(Test outcome negative) 7 10 7
TOP(Test outcome positive) 5 2 5
TP(True positive/hit) 3 1 3
TPR(Sensitivity, recall, hit rate, or true positive rate) 1.0 0.33333 0.5
Direct CM
>>> from pycm import *
>>> cm2 = ConfusionMatrix(matrix={"Class1": {"Class1": 1, "Class2": 2}, "Class2": {"Class1": 0, "Class2": 5}})
>>> cm2
pycm.ConfusionMatrix(classes: ['Class1', 'Class2'])
>>> cm2.classes
['Class1', 'Class2']
>>> cm2.print_matrix()
Predict Class1 Class2
Actual
Class1 1 2
Class2 0 5
>>> cm2.print_normalized_matrix()
Predict Class1 Class2
Actual
Class1 0.33333 0.66667
Class2 0.0 1.0
>>> cm2.stat(summary=True)
Overall Statistics :
ACC Macro 0.75
F1 Macro 0.66667
FPR Macro 0.33333
Kappa 0.38462
Overall ACC 0.75
PPV Macro 0.85714
SOA1(Landis & Koch) Fair
TPR Macro 0.66667
Zero-one Loss 2
Class Statistics :
Classes Class1 Class2
ACC(Accuracy) 0.75 0.75
AUC(Area under the ROC curve) 0.66667 0.66667
AUCI(AUC value interpretation) Fair Fair
F1(F1 score - harmonic mean of precision and sensitivity) 0.5 0.83333
FN(False negative/miss/type 2 error) 2 0
FP(False positive/type 1 error/false alarm) 0 2
FPR(Fall-out or false positive rate) 0.0 0.66667
N(Condition negative) 5 3
P(Condition positive or support) 3 5
POP(Population) 8 8
PPV(Precision or positive predictive value) 1.0 0.71429
TN(True negative/correct rejection) 5 1
TON(Test outcome negative) 7 1
TOP(Test outcome positive) 1 7
TP(True positive/hit) 1 5
TPR(Sensitivity, recall, hit rate, or true positive rate) 0.33333 1.0
matrix()andnormalized_matrix()renamed toprint_matrix()andprint_normalized_matrix()inversion 1.5
Activation threshold
threshold is added in version 0.9 for real value prediction.
For more information visit Example3
Load from file
file is added in version 0.9.5 in order to load saved confusion matrix with .obj format generated by save_obj method.
For more information visit Example4
Sample weights
sample_weight is added in version 1.2
For more information visit Example5
Transpose
transpose is added in version 1.2 in order to transpose input matrix (only in Direct CM mode)
Relabel
relabel method is added in version 1.5 in order to change ConfusionMatrix classnames.
>>> cm.relabel(mapping={0: "L1", 1: "L2", 2: "L3"})
>>> cm
pycm.ConfusionMatrix(classes: ['L1', 'L2', 'L3'])
Position
position method is added in version 2.8 in order to find the indexes of observations in predict_vector which made TP, TN, FP, FN.
>>> cm.position()
{0: {'FN': [], 'FP': [0, 7], 'TP': [1, 4, 9], 'TN': [2, 3, 5, 6, 8, 10, 11]}, 1: {'FN': [5, 10], 'FP': [3], 'TP': [6], 'TN': [0, 1, 2, 4, 7, 8, 9, 11]}, 2: {'FN': [0, 3, 7], 'FP': [5, 10], 'TP': [2, 8, 11], 'TN': [1, 4, 6, 9]}}
To array
to_array method is added in version 2.9 in order to returns the confusion matrix in the form of a NumPy array. This can be helpful to apply different operations over the confusion matrix for different purposes such as aggregation, normalization, and combination.
>>> cm.to_array()
array([[3, 0, 0],
[0, 1, 2],
[2, 1, 3]])
>>> cm.to_array(normalized=True)
array([[1. , 0. , 0. ],
[0. , 0.33333, 0.66667],
[0.33333, 0.16667, 0.5 ]])
>>> cm.to_array(normalized=True, one_vs_all=True, class_name="L1")
array([[1. , 0. ],
[0.22222, 0.77778]])
Combine
combine method is added in version 3.0 in order to merge two confusion matrices. This option will be useful in mini-batch learning.
>>> cm_combined = cm2.combine(cm3)
>>> cm_combined.print_matrix()
Predict Class1 Class2
Actual
Class1 2 4
Class2 0 10
Plot
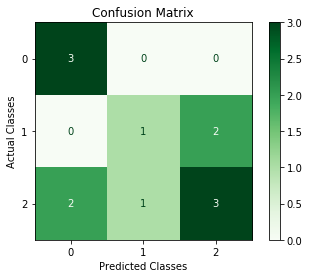
plot method is added in version 3.0 in order to plot a confusion matrix using Matplotlib or Seaborn.
>>> cm.plot()

>>> from matplotlib import pyplot as plt
>>> cm.plot(cmap=plt.cm.Greens, number_label=True, plot_lib="matplotlib")
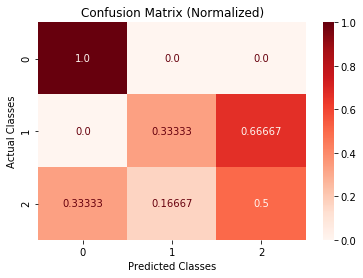
>>> cm.plot(cmap=plt.cm.Reds, normalized=True, number_label=True, plot_lib="seaborn")
ROC curve
ROCCurve, added in version 3.7, is devised to compute the Receiver Operating Characteristic (ROC) or simply ROC curve. In ROC curves, the Y axis represents the True Positive Rate, and the X axis represents the False Positive Rate. Thus, the ideal point is located at the top left of the curve, and a larger area under the curve represents better performance. ROC curve is a graphical representation of binary classifiers' performance. In PyCM, ROCCurve binarizes the output based on the "One vs. Rest" strategy to provide an extension of ROC for multi-class classifiers. Getting the actual labels vector, the target probability estimates of the positive classes, and the list of ordered labels of classes, this method is able to compute and plot TPR-FPR pairs for different discrimination thresholds and compute the area under the ROC curve.
>>> crv = ROCCurve(actual_vector=np.array([1, 1, 2, 2]), probs=np.array([[0.1, 0.9], [0.4, 0.6], [0.35, 0.65], [0.8, 0.2]]), classes=[2, 1])
>>> crv.thresholds
[0.1, 0.2, 0.35, 0.4, 0.6, 0.65, 0.8, 0.9]
>>> auc_trp = crv.area()
>>> auc_trp[1]
0.75
>>> auc_trp[2]
0.75
>>> optimal_thresholds = crv.optimal_thresholds()
>>> optimal_thresholds[1]
0.35
>>> optimal_thresholds[2]
0.2
ℹ️ Starting from version 4.5, the optimal_thresholds() method is available to calculate class-specific optimal cut-points using the "Closest to (0,1)" criterion. This method finds the threshold for each class that minimizes the Euclidean distance to the perfect classifier point.
Precision-Recall curve
PRCurve, added in version 3.7, is devised to compute the Precision-Recall curve in which the Y axis represents the Precision, and the X axis represents the Recall of a classifier. Thus, the ideal point is located at the top right of the curve, and a larger area under the curve represents better performance. Precision-Recall curve is a graphical representation of binary classifiers' performance. In PyCM, PRCurve binarizes the output based on the "One vs. Rest" strategy to provide an extension of this curve for multi-class classifiers. Getting the actual labels vector, the target probability estimates of the positive classes, and the list of ordered labels of classes, this method is able to compute and plot Precision-Recall pairs for different discrimination thresholds and compute the area under the curve.
>>> crv = PRCurve(actual_vector=np.array([1, 1, 2, 2]), probs=np.array([[0.1, 0.9], [0.4, 0.6], [0.35, 0.65], [0.8, 0.2]]), classes=[2, 1])
>>> crv.thresholds
[0.1, 0.2, 0.35, 0.4, 0.6, 0.65, 0.8, 0.9]
>>> auc_trp = crv.area()
>>> auc_trp[1]
0.29166666666666663
>>> auc_trp[2]
0.29166666666666663
Parameter recommender
This option has been added in version 1.9 to recommend the most related parameters considering the characteristics of the input dataset.
The suggested parameters are selected according to some characteristics of the input such as being balance/imbalance and binary/multi-class.
All suggestions can be categorized into three main groups: imbalanced dataset, binary classification for a balanced dataset, and multi-class classification for a balanced dataset.
The recommendation lists have been gathered according to the respective paper of each parameter and the capabilities which had been claimed by the paper.
>>> cm.imbalance
False
>>> cm.binary
False
>>> cm.recommended_list
['MCC', 'TPR Micro', 'ACC', 'PPV Macro', 'BCD', 'Overall MCC', 'Hamming Loss', 'TPR Macro', 'Zero-one Loss', 'ERR', 'PPV Micro', 'Overall ACC']
is_imbalanced parameter has been added in version 3.3, so the user can indicate whether the concerned dataset is imbalanced or not. As long as the user does not provide any information in this regard, the automatic detection algorithm will be used.
>>> cm = ConfusionMatrix(y_actu, y_pred, is_imbalanced=True)
>>> cm.imbalance
True
>>> cm = ConfusionMatrix(y_actu, y_pred, is_imbalanced=False)
>>> cm.imbalance
False
Compare
In version 2.0, a method for comparing several confusion matrices is introduced. This option is a combination of several overall and class-based benchmarks. Each of the benchmarks evaluates the performance of the classification algorithm from good to poor and give them a numeric score. The score of good and poor performances are 1 and 0, respectively.
After that, two scores are calculated for each confusion matrices, overall and class-based. The overall score is the average of the score of seven overall benchmarks which are Landis & Koch, Cramer, Matthews, Goodman-Kruskal's Lambda A, Goodman-Kruskal's Lambda B, Krippendorff's Alpha, and Pearson's C. In the same manner, the class-based score is the average of the score of six class-based benchmarks which are Positive Likelihood Ratio Interpretation, Negative Likelihood Ratio Interpretation, Discriminant Power Interpretation, AUC value Interpretation, Matthews Correlation Coefficient Interpretation and Yule's Q Interpretation. It should be noticed that if one of the benchmarks returns none for one of the classes, that benchmarks will be eliminated in total averaging. If the user sets weights for the classes, the averaging over the value of class-based benchmark scores will transform to a weighted average.
If the user sets the value of by_class boolean input True, the best confusion matrix is the one with the maximum class-based score. Otherwise, if a confusion matrix obtains the maximum of both overall and class-based scores, that will be reported as the best confusion matrix, but in any other case, the compared object doesn’t select the best confusion matrix.
>>> cm2 = ConfusionMatrix(matrix={0: {0: 2, 1: 50, 2: 6}, 1: {0: 5, 1: 50, 2: 3}, 2: {0: 1, 1: 7, 2: 50}})
>>> cm3 = ConfusionMatrix(matrix={0: {0: 50, 1: 2, 2: 6}, 1: {0: 50, 1: 5, 2: 3}, 2: {0: 1, 1: 55, 2: 2}})
>>> cp = Compare({"cm2": cm2, "cm3": cm3})
>>> print(cp)
Best : cm2
Rank Name Class-Score Overall-Score
1 cm2 0.50278 0.58095
2 cm3 0.33611 0.52857
>>> cp.best
pycm.ConfusionMatrix(classes: [0, 1, 2])
>>> cp.sorted
['cm2', 'cm3']
>>> cp.best_name
'cm2'
Multilabel confusion matrix
From version 4.0, MultiLabelCM has been added to calculate class-wise or sample-wise multilabel confusion matrices. In class-wise mode, confusion matrices are calculated for each class, and in sample-wise mode, they are generated per sample. All generated confusion matrices are binarized with a one-vs-rest transformation.
>>> mlcm = MultiLabelCM(actual_vector=[{"cat", "bird"}, {"dog"}], predict_vector=[{"cat"}, {"dog", "bird"}], classes=["cat", "dog", "bird"])
>>> mlcm.actual_vector_multihot
[[1, 0, 1], [0, 1, 0]]
>>> mlcm.predict_vector_multihot
[[1, 0, 0], [0, 1, 1]]
>>> mlcm.get_cm_by_class("cat").print_matrix()
Predict 0 1
Actual
0 1 0
1 0 1
>>> mlcm.get_cm_by_sample(0).print_matrix()
Predict 0 1
Actual
0 1 0
1 1 1
Online help
online_help function is added in version 1.1 in order to open each statistics definition in web browser
>>> from pycm import online_help
>>> online_help("J")
>>> online_help("SOA1(Landis & Koch)")
>>> online_help(2)
- List of items are available by calling
online_help()(without argument) - If PyCM website is not available, set
alt_link = True(new inversion 2.4)
Screen record
Try PyCM in your browser!
PyCM can be used online in interactive Jupyter Notebooks via the Binder or Colab services! Try it out now! :
- Check
ExamplesinDocumentfolder
Issues & bug reports
- Fill an issue and describe it. We'll check it ASAP!
- Please complete the issue template
- Discord : https://discord.com/invite/zqpU2b3J3f
- Website : https://www.pycm.io
- Mailing List : https://mail.python.org/mailman3/lists/pycm.python.org/
- Email : info@pycm.io
Acknowledgments
NLnet foundation has supported the PyCM project from version 4.3 to 4.7 through the NGI0 Commons Fund. This fund is set up by NLnet foundation with funding from the European Commission's Next Generation Internet program, administered by DG Communications Networks, Content, and Technology under grant agreement No 101135429.
NLnet foundation has supported the PyCM project from version 3.6 to 4.0 through the NGI Assure Fund. This fund is set up by NLnet foundation with funding from the European Commission's Next Generation Internet program, administered by DG Communications Networks, Content, and Technology under grant agreement No 957073.
Python Software Foundation (PSF) grants PyCM library partially for version 3.7. PSF is the organization behind Python. Their mission is to promote, protect, and advance the Python programming language and to support and facilitate the growth of a diverse and international community of Python programmers.
Some parts of the infrastructure for this project are supported by:
Cite
If you use PyCM in your research, we would appreciate citations to the following paper:
@article{Haghighi2018,
doi = {10.21105/joss.00729},
url = {https://doi.org/10.21105/joss.00729},
year = {2018},
month = {may},
publisher = {The Open Journal},
volume = {3},
number = {25},
pages = {729},
author = {Sepand Haghighi and Masoomeh Jasemi and Shaahin Hessabi and Alireza Zolanvari},
title = {{PyCM}: Multiclass confusion matrix library in Python},
journal = {Journal of Open Source Software}
}
Download PyCM.bib
| JOSS |  |
| Zenodo |  |
Show your support
Star this repo
Give a ⭐️ if this project helped you!
Donate to our project
If you do like our project and we hope that you do, can you please support us? Our project is not and is never going to be working for profit. We need the money just so we can continue doing what we do ;-) .
Changelog
All notable changes to this project will be documented in this file.
The format is based on Keep a Changelog and this project adheres to Semantic Versioning.
Unreleased
4.5 - 2025-10-15
Added
optimal_thresholdsmethod inROCCurveclass
Changed
- Python typing features added to modules
- Test system modified
4.4 - 2025-08-16
Added
print_timingsmethod- Test outcome positive rate
- Positive rate
run_report_benchmarkfunction
Changed
README.mdmodified- Test system modified
- Document modified
PRE_calcfunction renamed toproportion_calcPython 3.6support dropped
4.3 - 2025-04-04
Added
dissimilarity_matrixmethod
Changed
- HTML generator engine modified
README.mdmodified- Document modified
- String templates modified
Removed
html_initfunctionhtml_endfunction
4.2 - 2025-01-14
Added
- 5 new distance/similarity
- KuhnsIII
- KuhnsIV
- KuhnsV
- KuhnsVI
- KuhnsVII
Changed
- Test system modified
- PyPI badge in
README.md - GitHub actions are limited to the
devandmasterbranches AUTHORS.mdupdatedREADME.mdmodified- Document modified
4.1 - 2024-10-17
Added
- 5 new distance/similarity
- KoppenI
- KoppenII
- KuderRichardson
- KuhnsI
- KuhnsII
feature_request.ymltemplateconfig.ymlfor issue templateSECURITY.md
Changed
- Bug report template modified
thresholds_calcfunction updated__midpoint_numeric_integral__function updated__trapezoidal_numeric_integral__function updated- Diagrams updated
- Document modified
- Document build system updated
AUTHORS.mdupdatedREADME.mdmodified- Test system modified
Python 3.12added totest.ymlPython 3.13added totest.yml- Warning and error messages updated
pycm_util.pyrenamed toutils.pypycm_test.pyrenamed tobasic_test.pypycm_profile.pyrenamed toprofile.pypycm_param.pyrenamed toparams.pypycm_overall_func.pyrenamed tooverall_funcs.pypycm_output.pyrenamed tooutput.pypycm_obj.pyrenamed tocm.pypycm_multilabel_cm.pyrenamed tomultilabel_cm.pypycm_interpret.pyrenamed tointerpret.pypycm_handler.pyrenamed tohandlers.pypycm_error.pyrenamed toerrors.pypycm_distance.pyrenamed todistance.pypycm_curve.pyrenamed tocurve.pypycm_compare.pyrenamed tocompare.pypycm_class_func.pyrenamed toclass_funcs.pypycm_ci.pyrenamed toci.py
4.0 - 2023-06-07
Added
pycmMultiLabelErrorclassMultiLabelCMclassget_cm_by_classmethodget_cm_by_samplemethod__mlcm_vector_handler__function__mlcm_assign_classes__function__mlcm_vectors_filter__function__set_to_multihot__functiondeprecatedfunction
Changed
- Document modified
README.mdmodified- Example-4 modified
- Test system modified
- Python 3.5 support dropped
3.9 - 2023-05-01
Added
OVERALL_PARAMSdictionary__imbalancement_handler__functionvector_serializerfunction- NPV micro/macro
log_lossmethod- 23 new distance/similarity
- Dennis
- Digby
- Dispersion
- Doolittle
- Eyraud
- Fager & McGowan
- Faith
- Fleiss-Levin-Paik
- Forbes I
- Forbes II
- Fossum
- Gilbert & Wells
- Goodall
- Goodman & Kruskal's Lambda
- Goodman & Kruskal Lambda-r
- Guttman's Lambda A
- Guttman's Lambda B
- Hamann
- Harris & Lahey
- Hawkins & Dotson
- Kendall's Tau
- Kent & Foster I
- Kent & Foster II
Changed
metrics_offparameter added to ConfusionMatrix__init__methodCLASS_PARAMSchanged to a dictionary- Code style modified
sortparameter added torelabelmethod- Document modified
CONTRIBUTING.mdupdatedcodecovremoved fromdev-requirements.txt- Test system modified
3.8 - 2023-02-01
Added
distancemethod__contains__method__getitem__method- Goodman-Kruskal's Lambda A benchmark
- Goodman-Kruskal's Lambda B benchmark
- Krippendorff's Alpha benchmark
- Pearson's C benchmark
- 30 new distance/similarity
- AMPLE
- Anderberg's D
- Andres & Marzo's Delta
- Baroni-Urbani & Buser I
- Baroni-Urbani & Buser II
- Batagelj & Bren
- Baulieu I
- Baulieu II
- Baulieu III
- Baulieu IV
- Baulieu V
- Baulieu VI
- Baulieu VII
- Baulieu VIII
- Baulieu IX
- Baulieu X
- Baulieu XI
- Baulieu XII
- Baulieu XIII
- Baulieu XIV
- Baulieu XV
- Benini I
- Benini II
- Canberra
- Clement
- Consonni & Todeschini I
- Consonni & Todeschini II
- Consonni & Todeschini III
- Consonni & Todeschini IV
- Consonni & Todeschini V
Changed
relabelmethod sort bug fixedREADME.mdmodifiedCompareoverall benchmarks default weights updated- Document modified
- Test system modified
3.7 - 2022-12-15
Added
CurveclassROCCurveclassPRCurveclasspycmCurveErrorclass
Changed
CONTRIBUTING.mdupdatedmatrix_params_calcfunction optimizedREADME.mdmodified- Document modified
- Test system modified
Python 3.11added totest.yml
3.6 - 2022-08-17
Added
- Hamming distance
- Braun-Blanquet similarity
Changed
classesparameter added tomatrix_params_from_tablefunction- Matrices with
numpy.integerelements are now accepted - Arrays added to
matrixparameter accepting formats - Website changed to http://www.pycm.io
- Document modified
README.mdmodified
3.5 - 2022-04-27
Added
- Anaconda workflow
- Custom iterating setting
- Custom casting setting
Changed
plotmethod updatedclass_statisticsfunction modifiedoverall_statisticsfunction modifiedBCD_calcfunction modifiedCONTRIBUTING.mdupdatedCODE_OF_CONDUCT.mdupdated- Document modified
3.4 - 2022-01-26
Added
- Colab badge
- Discord badge
brier_scoremethod
Changed
J (Jaccard index)section inDocument.ipynbupdatedsave_objmethod updatedPython 3.10added totest.yml- Example-3 updated
- Docstrings of the functions updated
CONTRIBUTING.mdupdated
3.3 - 2021-10-27
Added
__compare_weight_handler__function
Changed
is_imbalancedparameter added to ConfusionMatrix__init__methodclass_benchmark_weightandoverall_benchmark_weightparameters added to Compare__init__methodstatistic_recommendfunction modified- Compare
weightparameter renamed toclass_weight - Document modified
- License updated
AUTHORS.mdupdatedREADME.mdmodified- Block diagrams updated
3.2 - 2021-08-11
Added
classes_filterfunction
Changed
classesparameter added tomatrix_params_calcfunctionclassesparameter added to__obj_vector_handler__functionclassesparameter added to ConfusionMatrix__init__methodnameparameter removed fromhtml_initfunctionshortenerparameter added tohtml_tablefunctionshortenerparameter added tosave_htmlmethod- Document modified
- HTML report modified
3.1 - 2021-03-11
Added
requirements-splitter.pysensitivity_indexmethod
Changed
- Test system modified
overall_statisticsfunction modified- HTML report modified
- Document modified
- References format updated
CONTRIBUTING.mdupdated
3.0 - 2020-10-26
Added
plot_test.pyaxes_genfunctionadd_number_labelfunctionplotmethodcombinemethodmatrix_combinefunction
Changed
- Document modified
README.mdmodified- Example-2 deprecated
- Example-7 deprecated
- Error messages modified
2.9 - 2020-09-23
Added
notebook_check.pyto_arraymethod__copy__methodcopymethod
Changed
averagemethod refactored
2.8 - 2020-07-09
Added
label_mapattributepositionsattributepositionmethod- Krippendorff's Alpha
- Aickin's Alpha
weighted_alphamethod
Changed
- Single class bug fixed
CLASS_NUMBER_ERRORerror type changed topycmMatrixErrorrelabelmethod bug fixed- Document modified
README.mdmodified
2.7 - 2020-05-11
Added
averagemethodweighted_averagemethodweighted_kappamethodpycmAverageErrorclass- Bangdiwala's B
- MATLAB examples
- Github action
Changed
- Document modified
README.mdmodifiedrelabelmethod bug fixedsparse_table_printfunction bug fixedmatrix_checkfunction bug fixed- Minor bug in
Compareclass fixed - Class names mismatch bug fixed
2.6 - 2020-03-25
Added
custom_rounderfunctioncomplementfunctionsparse_matrixattributesparse_normalized_matrixattribute- Net benefit (NB)
- Yule's Q interpretation (QI)
- Adjusted Rand index (ARI)
- TNR micro/macro
- FPR micro/macro
- FNR micro/macro
Changed
sparseparameter added toprint_matrix,print_normalized_matrixandsave_statmethodsheaderparameter added tosave_csvmethod- Handler functions moved to
pycm_handler.py - Error objects moved to
pycm_error.py - Verified tests references updated
- Verified tests moved to
verified_test.py - Test system modified
CONTRIBUTING.mdupdated- Namespace optimized
README.mdmodified- Document modified
print_normalized_matrixmethod modifiednormalized_table_calcfunction modifiedsetup.pymodified- summary mode updated
- Dockerfile updated
Python 3.8added to.travis.yamlandappveyor.yml
Removed
PC_PI_calcfunction
2.5 - 2019-10-16
Added
__version__variable- Individual classification success index (ICSI)
- Classification success index (CSI)
- Example-8 (Confidence interval)
install.shautopep8.sh- Dockerfile
CImethod (supported statistics :ACC,AUC,Overall ACC,Kappa,TPR,TNR,PPV,NPV,PLR,NLR,PRE)
Changed
test.shmoved to.travisfolder- Python 3.4 support dropped
- Python 2.7 support dropped
AUTHORS.mdupdatedsave_stat,save_csvandsave_htmlmethods Non-ASCII character bug fixed- Mixed type input vectors bug fixed
CONTRIBUTING.mdupdated- Example-3 updated
README.mdmodified- Document modified
CIattribute renamed toCI95kappa_se_calcfunction renamed tokappa_SE_calcse_calcfunction modified and renamed toSE_calc- CI/SE functions moved to
pycm_ci.py - Minor bug in
save_htmlmethod fixed
2.4 - 2019-07-31
Added
- Tversky index (TI)
- Area under the PR curve (AUPR)
FUNDING.yml
Changed
AUC_calcfunction modified- Document modified
summaryparameter added tosave_html,save_stat,save_csvandstatmethodssample_weightbug innumpyarray format fixed- Inputs manipulation bug fixed
- Test system modified
- Warning system modified
alt_linkparameter added tosave_htmlmethod andonline_helpfunctionCompareclass tests moved tocompare_test.py- Warning tests moved to
warning_test.py
2.3 - 2019-06-27
Added
- Adjusted F-score (AGF)
- Overlap coefficient (OC)
- Otsuka-Ochiai coefficient (OOC)
Changed
save_statandsave_vectorparameters added tosave_objmethod- Document modified
README.mdmodified- Parameters recommendation for imbalance dataset modified
- Minor bug in
Compareclass fixed pycm_helpfunction modified- Benchmarks color modified
2.2 - 2019-05-30
Added
- Negative likelihood ratio interpretation (NLRI)
- Cramer's benchmark (SOA5)
- Matthews correlation coefficient interpretation (MCCI)
- Matthews's benchmark (SOA6)
- F1 macro
- F1 micro
- Accuracy macro
Changed
Compareclass score calculation modified- Parameters recommendation for multi-class dataset modified
- Parameters recommendation for imbalance dataset modified
README.mdmodified- Document modified
- Logo updated
2.1 - 2019-05-06
Added
- Adjusted geometric mean (AGM)
- Yule's Q (Q)
Compareclass and parameters recommendation system block diagrams
Changed
- Document links bug fixed
- Document modified
2.0 - 2019-04-15
Added
- G-Mean (GM)
- Index of balanced accuracy (IBA)
- Optimized precision (OP)
- Pearson's C (C)
Compareclass- Parameters recommendation warning
ConfusionMatrixequal method
Changed
- Document modified
stat_printfunction bug fixedtable_printfunction bug fixedBetaparameter renamed tobeta(F_calcfunction &F_betamethod)- Parameters recommendation for imbalance dataset modified
normalizeparameter added tosave_htmlmethodpycm_func.pysplitted intopycm_class_func.pyandpycm_overall_func.pyvector_filter,vector_check,class_checkandmatrix_checkfunctions moved topycm_util.pyRACC_calcandRACCU_calcfunctions exception handler modified- Docstrings modified
1.9 - 2019-02-25
Added
- Automatic/Manual (AM)
- Bray-Curtis dissimilarity (BCD)
CODE_OF_CONDUCT.mdISSUE_TEMPLATE.mdPULL_REQUEST_TEMPLATE.mdCONTRIBUTING.md- X11 color names support for
save_htmlmethod - Parameters recommendation system
- Warning message for high dimension matrix print
- Interactive notebooks section (binder)
Changed
save_matrixandnormalizeparameters added tosave_csvmethodREADME.mdmodified- Document modified
ConfusionMatrix.__init__optimized- Document and examples output files moved to different folders
- Test system modified
relabelmethod bug fixed
1.8 - 2019-01-05
Added
- Lift score (LS)
version_check.py
Changed
colorparameter added tosave_htmlmethod- Error messages modified
- Document modified
- Website changed to http://www.pycm.ir
- Interpretation functions moved to
pycm_interpret.py - Utility functions moved to
pycm_util.py - Unnecessary
elseandelifremoved ==changed tois
1.7 - 2018-12-18
Added
- Gini index (GI)
- Example-7
pycm_profile.py
Changed
class_nameparameter added tostat,save_stat,save_csvandsave_htmlmethodsoverall_paramandclass_paramparameters empty list bug fixedmatrix_params_calc,matrix_params_from_tableandvector_filterfunctions optimizedoverall_MCC_calc,CEN_misclassification_calcandconvex_combinationfunctions optimized- Document modified
1.6 - 2018-12-06
Added
- AUC value interpretation (AUCI)
- Example-6
- Anaconda cloud package
Changed
overall_paramandclass_paramparameters added tostat,save_statandsave_htmlmethodsclass_paramparameter added tosave_csvmethod_removed from overall statistics namesREADME.mdmodified- Document modified
1.5 - 2018-11-26
Added
- Relative classifier information (RCI)
- Discriminator power (DP)
- Youden's index (Y)
- Discriminant power interpretation (DPI)
- Positive likelihood ratio interpretation (PLRI)
__len__methodrelabelmethod__class_stat_init__function__overall_stat_init__functionmatrixattribute as dictnormalized_matrixattribute as dictnormalized_tableattribute as dict
Changed
README.mdmodified- Document modified
LR+renamed toPLRLR-renamed toNLRnormalized_matrixmethod renamed toprint_normalized_matrixmatrixmethod renamed toprint_matrixentropy_calcfixedcross_entropy_calcfixedconditional_entropy_calcfixedprint_tablebug for large numbers fixed- JSON key bug in
save_objfixed transposebug insave_objfixedPython 3.7added to.travis.yamlandappveyor.yml
1.4 - 2018-11-12
Added
- Area under curve (AUC)
- AUNU
- AUNP
- Class balance accuracy (CBA)
- Global performance index (RR)
- Overall MCC
- Distance index (dInd)
- Similarity index (sInd)
one_vs_alldev-requirements.txt
Changed
README.mdmodified- Document modified
save_statmodifiedrequirements.txtmodified
1.3 - 2018-10-10
Added
- Confusion entropy (CEN)
- Overall confusion entropy (Overall CEN)
- Modified confusion entropy (MCEN)
- Overall modified confusion entropy (Overall MCEN)
- Information score (IS)
Changed
README.mdmodified
1.2 - 2018-10-01
Added
- No information rate (NIR)
- P-Value
sample_weighttranspose
Changed
README.mdmodified- Key error in some parameters fixed
OSXenv added to.travis.yml
1.1 - 2018-09-08
Added
- Zero-one loss
- Support
online_helpfunction
Changed
README.mdmodifiedhtml_tablefunction modifiedtable_printfunction modifiednormalized_table_printfunction modified
1.0 - 2018-08-30
Added
- Hamming loss
Changed
README.mdmodified
0.9.5 - 2018-07-08
Added
- Obj load
- Obj save
- Example-4
Changed
README.mdmodified- Block diagram updated
0.9 - 2018-06-28
Added
- Activation threshold
- Example-3
- Jaccard index
- Overall Jaccard index
Changed
README.mdmodifiedsetup.pymodified
0.8.6 - 2018-05-31
Added
- Example section in document
- Python 2.7 CI
- JOSS paper pdf
Changed
- Cite section
- ConfusionMatrix docstring
- round function changed to numpy.around
README.mdmodified
0.8.5 - 2018-05-21
Added
- Example-1 (Comparison of three different classifiers)
- Example-2 (How to plot via matplotlib)
- JOSS paper
- ConfusionMatrix docstring
Changed
- Table size in HTML report
- Test system
README.mdmodified
0.8.1 - 2018-03-22
Added
- Goodman and Kruskal's lambda B
- Goodman and Kruskal's lambda A
- Cross entropy
- Conditional entropy
- Joint entropy
- Reference entropy
- Response entropy
- Kullback-Liebler divergence
- Direct ConfusionMatrix
- Kappa unbiased
- Kappa no prevalence
- Random accuracy unbiased
pycmVectorErrorclasspycmMatrixErrorclass- Mutual information
- Support
numpyarrays
Changed
- Notebook file updated
Removed
pycmErrorclass
0.7 - 2018-02-26
Added
- Cramer's V
- 95% confidence interval
- Chi-Squared
- Phi-Squared
- Chi-Squared DF
- Standard error
- Kappa standard error
- Kappa 95% confidence interval
- Cicchetti benchmark
Changed
- Overall statistics color in HTML report
- Parameters description link in HTML report
0.6 - 2018-02-21
Added
- CSV report
- Changelog
- Output files
digitparameter toConfusionMatrixobject
Changed
- Confusion matrix color in HTML report
- Parameters description link in HTML report
- Capitalize descriptions
0.5 - 2018-02-17
Added
- Scott's pi
- Gwet's AC1
- Bennett S score
- HTML report
0.4 - 2018-02-05
Added
- TPR micro/macro
- PPV micro/macro
- Overall RACC
- Error rate (ERR)
- FBeta score
- F0.5
- F2
- Fleiss benchmark
- Altman benchmark
- Output file(.pycm)
Changed
- Class with zero item
- Normalized matrix
Removed
- Kappa and SOA for each class
0.3 - 2018-01-27
Added
- Kappa
- Random accuracy
- Landis and Koch benchmark
overall_stat
0.2 - 2018-01-24
Added
- Population
- Condition positive
- Condition negative
- Test outcome positive
- Test outcome negative
- Prevalence
- G-measure
- Matrix method
- Normalized matrix method
- Params method
Changed
statistic_resulttoclass_statparamstostat
0.1 - 2018-01-22
Added
- ACC
- BM
- DOR
- F1-Score
- FDR
- FNR
- FOR
- FPR
- LR+
- LR-
- MCC
- MK
- NPV
- PPV
- TNR
- TPR
- documents and
README.md
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pycm-4.5.tar.gz.
File metadata
- Download URL: pycm-4.5.tar.gz
- Upload date:
- Size: 920.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9454ac2068967d8f1d00c40b530d455ddb6d786f04d4abe87ab0b15ab47e0893
|
|
| MD5 |
2f1b324d581ad563d5b5fdd2e448a1c8
|
|
| BLAKE2b-256 |
dca8887d0ff5a29dd2c1599ddeee70160cfeae3c62fdc07767a972bef86961d0
|
File details
Details for the file pycm-4.5-py3-none-any.whl.
File metadata
- Download URL: pycm-4.5-py3-none-any.whl
- Upload date:
- Size: 73.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
1f1f204710e4612bc6f08c5abb861cbb515c8931ee84a406320fd04920cac77a
|
|
| MD5 |
c5880f1618b157140e238b1184f04bd3
|
|
| BLAKE2b-256 |
5bba0e8f2833710934d63a04fb1a5774cc134e05a3289940fe1f43c496d07a50
|