Multi-class confusion matrix library in Python
Project description
Table of contents
- Overview
- Installation
- Usage
- Document
- Try PyCM in Your Browser
- Issues & Bug Reports
- Todo
- Outputs
- Dependencies
- Contribution
- References
- Cite
- Authors
- License
- Donate
- Changelog
- Code of Conduct
Overview
PyCM is a multi-class confusion matrix library written in Python that supports both input data vectors and direct matrix, and a proper tool for post-classification model evaluation that supports most classes and overall statistics parameters. PyCM is the swiss-army knife of confusion matrices, targeted mainly at data scientists that need a broad array of metrics for predictive models and an accurate evaluation of large variety of classifiers.
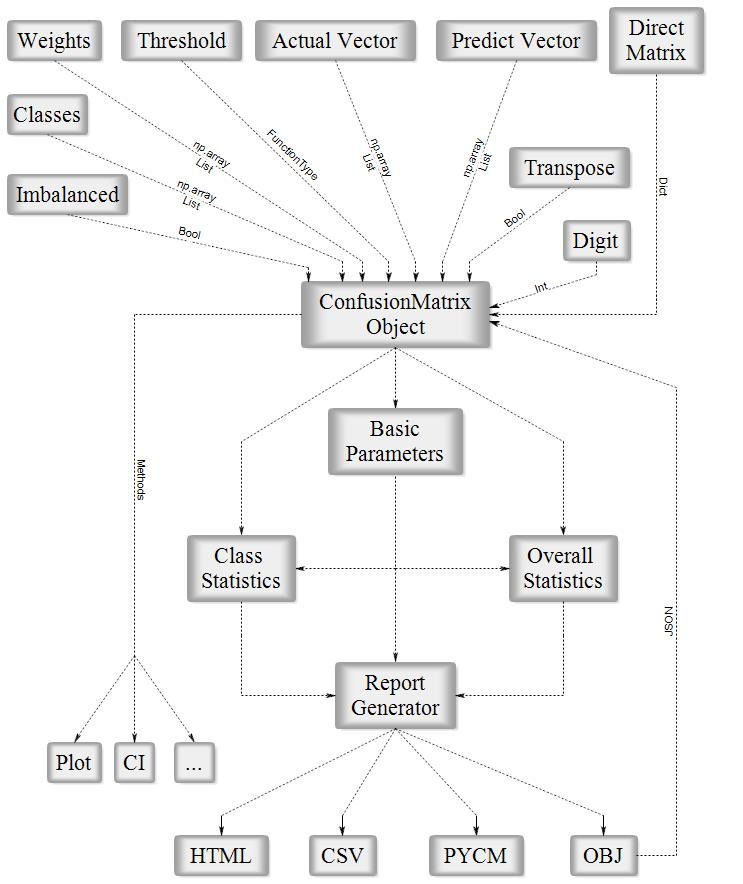
Fig1. ConfusionMatrix Block Diagram
| Open Hub |  |
| PyPI Counter |  |
| Github Stars |  |
| Branch | master | dev |
| Travis |  |
 |
| AppVeyor |  |
 |
| Code Quality |  |
 |
 |
Installation
Source code
- Download Version 2.3 or Latest Source
- Run
pip install -r requirements.txtorpip3 install -r requirements.txt(Need root access) - Run
python3 setup.py installorpython setup.py install(Need root access)
PyPI
- Check Python Packaging User Guide
- Run
pip install pycm==2.3orpip3 install pycm==2.3(Need root access)
Conda
- Check Conda Managing Package
conda install -c sepandhaghighi pycm(Need root access)
Easy install
- Run
easy_install --upgrade pycm(Need root access)
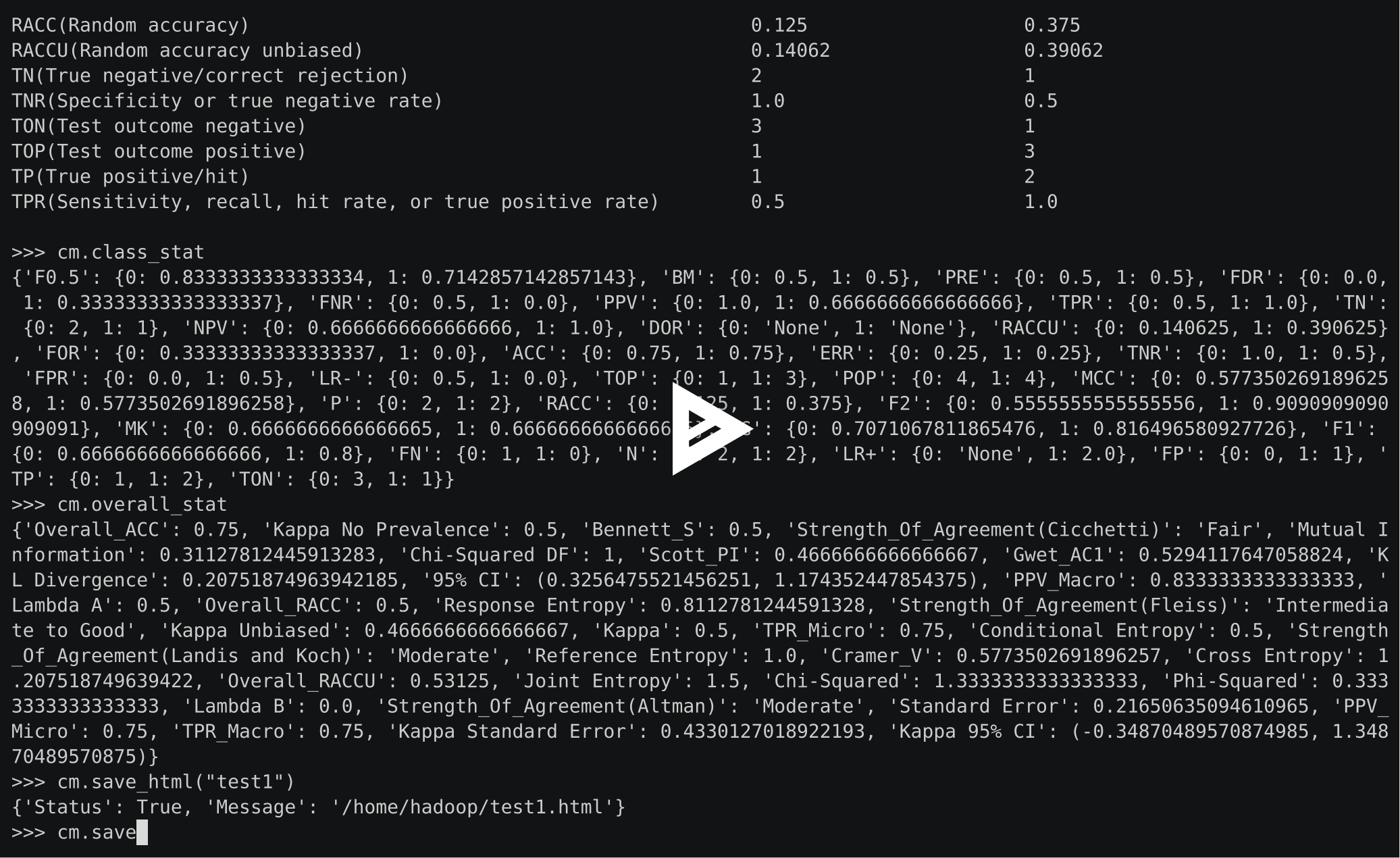
Usage
From vector
>>> from pycm import *
>>> y_actu = [2, 0, 2, 2, 0, 1, 1, 2, 2, 0, 1, 2] # or y_actu = numpy.array([2, 0, 2, 2, 0, 1, 1, 2, 2, 0, 1, 2])
>>> y_pred = [0, 0, 2, 1, 0, 2, 1, 0, 2, 0, 2, 2] # or y_pred = numpy.array([0, 0, 2, 1, 0, 2, 1, 0, 2, 0, 2, 2])
>>> cm = ConfusionMatrix(actual_vector=y_actu, predict_vector=y_pred) # Create CM From Data
>>> cm.classes
[0, 1, 2]
>>> cm.table
{0: {0: 3, 1: 0, 2: 0}, 1: {0: 0, 1: 1, 2: 2}, 2: {0: 2, 1: 1, 2: 3}}
>>> print(cm)
Predict 0 1 2
Actual
0 3 0 0
1 0 1 2
2 2 1 3
Overall Statistics :
95% CI (0.30439,0.86228)
ACC Macro 0.72222
AUNP 0.66667
AUNU 0.69444
Bennett S 0.375
CBA 0.47778
Chi-Squared 6.6
Chi-Squared DF 4
Conditional Entropy 0.95915
Cramer V 0.5244
Cross Entropy 1.59352
F1 Macro 0.56515
F1 Micro 0.58333
Gwet AC1 0.38931
Hamming Loss 0.41667
Joint Entropy 2.45915
KL Divergence 0.09352
Kappa 0.35484
Kappa 95% CI (-0.07708,0.78675)
Kappa No Prevalence 0.16667
Kappa Standard Error 0.22036
Kappa Unbiased 0.34426
Lambda A 0.16667
Lambda B 0.42857
Mutual Information 0.52421
NIR 0.5
Overall ACC 0.58333
Overall CEN 0.46381
Overall J (1.225,0.40833)
Overall MCC 0.36667
Overall MCEN 0.51894
Overall RACC 0.35417
Overall RACCU 0.36458
P-Value 0.38721
PPV Macro 0.56667
PPV Micro 0.58333
Pearson C 0.59568
Phi-Squared 0.55
RCI 0.34947
RR 4.0
Reference Entropy 1.5
Response Entropy 1.48336
SOA1(Landis & Koch) Fair
SOA2(Fleiss) Poor
SOA3(Altman) Fair
SOA4(Cicchetti) Poor
SOA5(Cramer) Relatively Strong
SOA6(Matthews) Weak
Scott PI 0.34426
Standard Error 0.14232
TPR Macro 0.61111
TPR Micro 0.58333
Zero-one Loss 5
Class Statistics :
Classes 0 1 2
ACC(Accuracy) 0.83333 0.75 0.58333
AGF(Adjusted F-score) 0.9136 0.53995 0.5516
AGM(Adjusted geometric mean) 0.83729 0.692 0.60712
AM(Difference between automatic and manual classification) 2 -1 -1
AUC(Area under the roc curve) 0.88889 0.61111 0.58333
AUCI(AUC value interpretation) Very Good Fair Poor
BCD(Bray-Curtis dissimilarity) 0.08333 0.04167 0.04167
BM(Informedness or bookmaker informedness) 0.77778 0.22222 0.16667
CEN(Confusion entropy) 0.25 0.49658 0.60442
DOR(Diagnostic odds ratio) None 4.0 2.0
DP(Discriminant power) None 0.33193 0.16597
DPI(Discriminant power interpretation) None Poor Poor
ERR(Error rate) 0.16667 0.25 0.41667
F0.5(F0.5 score) 0.65217 0.45455 0.57692
F1(F1 score - harmonic mean of precision and sensitivity) 0.75 0.4 0.54545
F2(F2 score) 0.88235 0.35714 0.51724
FDR(False discovery rate) 0.4 0.5 0.4
FN(False negative/miss/type 2 error) 0 2 3
FNR(Miss rate or false negative rate) 0.0 0.66667 0.5
FOR(False omission rate) 0.0 0.2 0.42857
FP(False positive/type 1 error/false alarm) 2 1 2
FPR(Fall-out or false positive rate) 0.22222 0.11111 0.33333
G(G-measure geometric mean of precision and sensitivity) 0.7746 0.40825 0.54772
GI(Gini index) 0.77778 0.22222 0.16667
GM(G-mean geometric mean of specificity and sensitivity) 0.88192 0.54433 0.57735
IBA(Index of balanced accuracy) 0.95062 0.13169 0.27778
IS(Information score) 1.26303 1.0 0.26303
J(Jaccard index) 0.6 0.25 0.375
LS(Lift score) 2.4 2.0 1.2
MCC(Matthews correlation coefficient) 0.68313 0.2582 0.16903
MCCI(Matthews correlation coefficient interpretation) Moderate Negligible Negligible
MCEN(Modified confusion entropy) 0.26439 0.5 0.6875
MK(Markedness) 0.6 0.3 0.17143
N(Condition negative) 9 9 6
NLR(Negative likelihood ratio) 0.0 0.75 0.75
NLRI(Negative likelihood ratio interpretation) Good Negligible Negligible
NPV(Negative predictive value) 1.0 0.8 0.57143
OC(Overlap coefficient) 1.0 0.5 0.6
OOC(Otsuka-Ochiai coefficient) 0.7746 0.40825 0.54772
OP(Optimized precision) 0.70833 0.29545 0.44048
P(Condition positive or support) 3 3 6
PLR(Positive likelihood ratio) 4.5 3.0 1.5
PLRI(Positive likelihood ratio interpretation) Poor Poor Poor
POP(Population) 12 12 12
PPV(Precision or positive predictive value) 0.6 0.5 0.6
PRE(Prevalence) 0.25 0.25 0.5
Q(Yule Q - coefficient of colligation) None 0.6 0.33333
RACC(Random accuracy) 0.10417 0.04167 0.20833
RACCU(Random accuracy unbiased) 0.11111 0.0434 0.21007
TN(True negative/correct rejection) 7 8 4
TNR(Specificity or true negative rate) 0.77778 0.88889 0.66667
TON(Test outcome negative) 7 10 7
TOP(Test outcome positive) 5 2 5
TP(True positive/hit) 3 1 3
TPR(Sensitivity, recall, hit rate, or true positive rate) 1.0 0.33333 0.5
Y(Youden index) 0.77778 0.22222 0.16667
dInd(Distance index) 0.22222 0.67586 0.60093
sInd(Similarity index) 0.84287 0.52209 0.57508
>>> cm.print_matrix()
Predict 0 1 2
Actual
0 3 0 0
1 0 1 2
2 2 1 3
>>> cm.print_normalized_matrix()
Predict 0 1 2
Actual
0 1.0 0.0 0.0
1 0.0 0.33333 0.66667
2 0.33333 0.16667 0.5
>>> cm.print_matrix(one_vs_all=True,class_name=0) # One-Vs-All, new in version 1.4
Predict 0 ~
Actual
0 3 0
~ 2 7
Direct CM
>>> from pycm import *
>>> cm2 = ConfusionMatrix(matrix={"Class1": {"Class1": 1, "Class2":2}, "Class2": {"Class1": 0, "Class2": 5}}) # Create CM Directly
>>> cm2
pycm.ConfusionMatrix(classes: ['Class1', 'Class2'])
>>> print(cm2)
Predict Class1 Class2
Actual
Class1 1 2
Class2 0 5
Overall Statistics :
95% CI (0.44994,1.05006)
ACC Macro 0.75
AUNP 0.66667
AUNU 0.66667
Bennett S 0.5
CBA 0.52381
Chi-Squared 1.90476
Chi-Squared DF 1
Conditional Entropy 0.34436
Cramer V 0.48795
Cross Entropy 1.2454
F1 Macro 0.66667
F1 Micro 0.75
Gwet AC1 0.6
Hamming Loss 0.25
Joint Entropy 1.29879
KL Divergence 0.29097
Kappa 0.38462
Kappa 95% CI (-0.354,1.12323)
Kappa No Prevalence 0.5
Kappa Standard Error 0.37684
Kappa Unbiased 0.33333
Lambda A 0.33333
Lambda B 0.0
Mutual Information 0.1992
NIR 0.625
Overall ACC 0.75
Overall CEN 0.44812
Overall J (1.04762,0.52381)
Overall MCC 0.48795
Overall MCEN 0.29904
Overall RACC 0.59375
Overall RACCU 0.625
P-Value 0.36974
PPV Macro 0.85714
PPV Micro 0.75
Pearson C 0.43853
Phi-Squared 0.2381
RCI 0.20871
RR 4.0
Reference Entropy 0.95443
Response Entropy 0.54356
SOA1(Landis & Koch) Fair
SOA2(Fleiss) Poor
SOA3(Altman) Fair
SOA4(Cicchetti) Poor
SOA5(Cramer) Relatively Strong
SOA6(Matthews) Weak
Scott PI 0.33333
Standard Error 0.15309
TPR Macro 0.66667
TPR Micro 0.75
Zero-one Loss 2
Class Statistics :
Classes Class1 Class2
ACC(Accuracy) 0.75 0.75
AGF(Adjusted F-score) 0.53979 0.81325
AGM(Adjusted geometric mean) 0.73991 0.5108
AM(Difference between automatic and manual classification) -2 2
AUC(Area under the roc curve) 0.66667 0.66667
AUCI(AUC value interpretation) Fair Fair
BCD(Bray-Curtis dissimilarity) 0.125 0.125
BM(Informedness or bookmaker informedness) 0.33333 0.33333
CEN(Confusion entropy) 0.5 0.43083
DOR(Diagnostic odds ratio) None None
DP(Discriminant power) None None
DPI(Discriminant power interpretation) None None
ERR(Error rate) 0.25 0.25
F0.5(F0.5 score) 0.71429 0.75758
F1(F1 score - harmonic mean of precision and sensitivity) 0.5 0.83333
F2(F2 score) 0.38462 0.92593
FDR(False discovery rate) 0.0 0.28571
FN(False negative/miss/type 2 error) 2 0
FNR(Miss rate or false negative rate) 0.66667 0.0
FOR(False omission rate) 0.28571 0.0
FP(False positive/type 1 error/false alarm) 0 2
FPR(Fall-out or false positive rate) 0.0 0.66667
G(G-measure geometric mean of precision and sensitivity) 0.57735 0.84515
GI(Gini index) 0.33333 0.33333
GM(G-mean geometric mean of specificity and sensitivity) 0.57735 0.57735
IBA(Index of balanced accuracy) 0.11111 0.55556
IS(Information score) 1.41504 0.19265
J(Jaccard index) 0.33333 0.71429
LS(Lift score) 2.66667 1.14286
MCC(Matthews correlation coefficient) 0.48795 0.48795
MCCI(Matthews correlation coefficient interpretation) Weak Weak
MCEN(Modified confusion entropy) 0.38998 0.51639
MK(Markedness) 0.71429 0.71429
N(Condition negative) 5 3
NLR(Negative likelihood ratio) 0.66667 0.0
NLRI(Negative likelihood ratio interpretation) Negligible Good
NPV(Negative predictive value) 0.71429 1.0
OC(Overlap coefficient) 1.0 1.0
OOC(Otsuka-Ochiai coefficient) 0.57735 0.84515
OP(Optimized precision) 0.25 0.25
P(Condition positive or support) 3 5
PLR(Positive likelihood ratio) None 1.5
PLRI(Positive likelihood ratio interpretation) None Poor
POP(Population) 8 8
PPV(Precision or positive predictive value) 1.0 0.71429
PRE(Prevalence) 0.375 0.625
Q(Yule Q - coefficient of colligation) None None
RACC(Random accuracy) 0.04688 0.54688
RACCU(Random accuracy unbiased) 0.0625 0.5625
TN(True negative/correct rejection) 5 1
TNR(Specificity or true negative rate) 1.0 0.33333
TON(Test outcome negative) 7 1
TOP(Test outcome positive) 1 7
TP(True positive/hit) 1 5
TPR(Sensitivity, recall, hit rate, or true positive rate) 0.33333 1.0
Y(Youden index) 0.33333 0.33333
dInd(Distance index) 0.66667 0.66667
sInd(Similarity index) 0.5286 0.5286
>>> cm3 = ConfusionMatrix(matrix={"Class1": {"Class1": 1, "Class2":0}, "Class2": {"Class1": 2, "Class2": 5}},transpose=True) # Transpose Matrix
>>> cm3.print_matrix()
Predict Class1 Class2
Actual
Class1 1 2
Class2 0 5
matrix()andnormalized_matrix()renamed toprint_matrix()andprint_normalized_matrix()inversion 1.5
Activation threshold
threshold is added in version 0.9 for real value prediction.
For more information visit Example3
Load from file
file is added in version 0.9.5 in order to load saved confusion matrix with .obj format generated by save_obj method.
For more information visit Example4
Sample weights
sample_weight is added in version 1.2
For more information visit Example5
Transpose
transpose is added in version 1.2 in order to transpose input matrix (only in Direct CM mode)
Relabel
relabel method is added in version 1.5 in order to change ConfusionMatrix classnames.
>>> cm.relabel(mapping={0:"L1",1:"L2",2:"L3"})
>>> cm
pycm.ConfusionMatrix(classes: ['L1', 'L2', 'L3'])
Online help
online_help function is added in version 1.1 in order to open each statistics definition in web browser
>>> from pycm import online_help
>>> online_help("J")
>>> online_help("SOA1(Landis & Koch)")
>>> online_help(2)
- List of items are available by calling
online_help()(without argument)
Parameter recommender
This option has been added in version 1.9 in order to recommend most related parameters considering the characteristics of the input dataset. The characteristics according to which the parameters are suggested are balance/imbalance and binary/multiclass. All suggestions can be categorized into three main groups: imbalanced dataset, binary classification for a balanced dataset, and multi-class classification for a balanced dataset. The recommendation lists have been gathered according to the respective paper of each parameter and the capabilities which had been claimed by the paper.
>>> cm.imbalance
False
>>> cm.binary
False
>>> cm.recommended_list
['MCC', 'TPR Micro', 'ACC', 'PPV Macro', 'BCD', 'Overall MCC', 'Hamming Loss', 'TPR Macro', 'Zero-one Loss', 'ERR', 'PPV Micro', 'Overall ACC']
Comapre
In version 2.0 a method for comparing several confusion matrices is introduced. This option is a combination of several overall and class-based benchmarks. Each of the benchmarks evaluates the performance of the classification algorithm from good to poor and give them a numeric score. The score of good performance is 1 and for the poor performance is 0.
After that, two scores are calculated for each confusion matrices, overall and class based. The overall score is the average of the score of six overall benchmarks which are Landis & Koch, Fleiss, Altman, Cicchetti, Cramer, and Matthews. And with a same manner, the class based score is the average of the score of five class-based benchmarks which are Positive Likelihood Ratio Interpretation, Negative Likelihood Ratio Interpretation, Discriminant Power Interpretation, AUC value Interpretation, and Matthews Correlation Coefficient Interpretation. It should be notice that if one of the benchmarks returns none for one of the classes, that benchmarks will be eliminate in total averaging. If user set weights for the classes, the averaging over the value of class-based benchmark scores will transform to a weighted average.
If the user set the value of by_class boolean input True, the best confusion matrix is the one with the maximum class-based score. Otherwise, if a confusion matrix obtain the maximum of the both overall and class-based score, that will be the reported as the best confusion matrix but in any other cases the compare object doesn’t select best confusion matrix.
>>> cm2 = ConfusionMatrix(matrix={0:{0:2,1:50,2:6},1:{0:5,1:50,2:3},2:{0:1,1:7,2:50}})
>>> cm3 = ConfusionMatrix(matrix={0:{0:50,1:2,2:6},1:{0:50,1:5,2:3},2:{0:1,1:55,2:2}})
>>> cp = Compare({"cm2":cm2,"cm3":cm3})
>>> print(cp)
Best : cm2
Rank Name Class-Score Overall-Score
1 cm2 4.15 1.48333
2 cm3 2.75 0.95
>>> cp.best
pycm.ConfusionMatrix(classes: [0, 1, 2])
>>> cp.sorted
['cm2', 'cm3']
>>> cp.best_name
'cm2'
Acceptable data types
ConfusionMatrix
actual_vector: pythonlistor numpyarrayof any stringable objectspredict_vector: pythonlistor numpyarrayof any stringable objectsmatrix:dictdigit:intthreshold:FunctionType (function or lambda)file:File objectsample_weight: pythonlistor numpyarrayof numberstranspose:bool
- Run
help(ConfusionMatrix)forConfusionMatrixobject details
Compare
cm_dict: pythondictofConfusionMatrixobject (str:ConfusionMatrix)by_class:boolweight: pythondictof class weights (class_name:float)digit:int
- Run
help(Compare)forCompareobject details
For more information visit here
Try PyCM in your browser!
PyCM can be used online in interactive Jupyter Notebooks via the Binder service! Try it out now! :
- Check
ExamplesinDocumentfolder
Issues & bug reports
Just fill an issue and describe it. We'll check it ASAP! or send an email to info@pycm.ir.
- Please complete the issue template
Outputs
Dependencies
| master | dev |
 |
 |
References
1- J. R. Landis, G. G. Koch, “The measurement of observer agreement for categorical data. Biometrics,” in International Biometric Society, pp. 159–174, 1977.
2- D. M. W. Powers, “Evaluation: from precision, recall and f-measure to roc, informedness, markedness & correlation,” in Journal of Machine Learning Technologies, pp.37-63, 2011.
3- C. Sammut, G. Webb, “Encyclopedia of Machine Learning” in Springer, 2011.
4- J. L. Fleiss, “Measuring nominal scale agreement among many raters,” in Psychological Bulletin, pp. 378-382, 1971.
5- D.G. Altman, “Practical Statistics for Medical Research,” in Chapman and Hall, 1990.
6- K. L. Gwet, “Computing inter-rater reliability and its variance in the presence of high agreement,” in The British Journal of Mathematical and Statistical Psychology, pp. 29–48, 2008.”
7- W. A. Scott, “Reliability of content analysis: The case of nominal scaling,” in Public Opinion Quarterly, pp. 321–325, 1955.
8- E. M. Bennett, R. Alpert, and A. C. Goldstein, “Communication through limited response questioning,” in The Public Opinion Quarterly, pp. 303–308, 1954.
9- D. V. Cicchetti, "Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology," in Psychological Assessment, pp. 284–290, 1994.
10- R.B. Davies, "Algorithm AS155: The Distributions of a Linear Combination of χ2 Random Variables," in Journal of the Royal Statistical Society, pp. 323–333, 1980.
11- S. Kullback, R. A. Leibler "On information and sufficiency," in Annals of Mathematical Statistics, pp. 79–86, 1951.
12- L. A. Goodman, W. H. Kruskal, "Measures of Association for Cross Classifications, IV: Simplification of Asymptotic Variances," in Journal of the American Statistical Association, pp. 415–421, 1972.
13- L. A. Goodman, W. H. Kruskal, "Measures of Association for Cross Classifications III: Approximate Sampling Theory," in Journal of the American Statistical Association, pp. 310–364, 1963.
14- T. Byrt, J. Bishop and J. B. Carlin, “Bias, prevalence, and kappa,” in Journal of Clinical Epidemiology pp. 423-429, 1993.
15- M. Shepperd, D. Bowes, and T. Hall, “Researcher Bias: The Use of Machine Learning in Software Defect Prediction,” in IEEE Transactions on Software Engineering, pp. 603-616, 2014.
16- X. Deng, Q. Liu, Y. Deng, and S. Mahadevan, “An improved method to construct basic probability assignment based on the confusion matrix for classification problem, ” in Information Sciences, pp.250-261, 2016.
17- J.-M. Wei, X.-J. Yuan, Q.-H. Hu, and S.-Q. J. E. S. w. A. Wang, "A novel measure for evaluating classifiers," in Expert Systems with Applications, pp. 3799-3809, 2010.
18- I. Kononenko and I. J. M. L. Bratko, "Information-based evaluation criterion for classifier's performance," in Machine Learning, pp. 67-80, 1991.
19- R. Delgado and J. D. Núñez-González, "Enhancing Confusion Entropy as Measure for Evaluating Classifiers," in The 13th International Conference on Soft Computing Models in Industrial and Environmental Applications, pp. 79-89, 2018: Springer.
20- J. J. C. b. Gorodkin and chemistry, "Comparing two K-category assignments by a K-category correlation coefficient," in Computational Biology and chemistry, pp. 367-374, 2004.
21- C. O. Freitas, J. M. De Carvalho, J. Oliveira, S. B. Aires, and R. Sabourin, "Confusion matrix disagreement for multiple classifiers," in Iberoamerican Congress on Pattern Recognition, pp. 387-396, 2007.
22- P. Branco, L. Torgo, and R. P. Ribeiro, "Relevance-based evaluation metrics for multi-class imbalanced domains," in Pacific-Asia Conference on Knowledge Discovery and Data Mining, pp. 698-710, 2017. Springer.
23- D. Ballabio, F. Grisoni, R. J. C. Todeschini, and I. L. Systems, "Multivariate comparison of classification performance measures," in Chemometrics and Intelligent Laboratory Systems, pp. 33-44, 2018.
24- J. J. E. Cohen and p. measurement, "A coefficient of agreement for nominal scales," in Educational and Psychological Measurement, pp. 37-46, 1960.
25- S. Siegel, "Nonparametric statistics for the behavioral sciences," in New York : McGraw-Hill, 1956.
26- H. Cramér, "Mathematical methods of statistics (PMS-9),"in Princeton university press, 2016.
27- B. W. J. B. e. B. A.-P. S. Matthews, "Comparison of the predicted and observed secondary structure of T4 phage lysozyme," in Biochimica et Biophysica Acta (BBA) - Protein Structure, pp. 442-451, 1975.
28- J. A. J. S. Swets, "The relative operating characteristic in psychology: a technique for isolating effects of response bias finds wide use in the study of perception and cognition," in American Association for the Advancement of Science, pp. 990-1000, 1973.
29- P. J. B. S. V. S. N. Jaccard, "Étude comparative de la distribution florale dans une portion des Alpes et des Jura," in Bulletin de la Société vaudoise des sciences naturelles, pp. 547-579, 1901.
30- T. M. Cover and J. A. Thomas, "Elements of information theory," in John Wiley & Sons, 2012.
31- E. S. Keeping, "Introduction to statistical inference," in Courier Corporation, 1995.
32- V. Sindhwani, P. Bhattacharya, and S. Rakshit, "Information theoretic feature crediting in multiclass support vector machines," in Proceedings of the 2001 SIAM International Conference on Data Mining, pp. 1-18, 2001.
33- M. Bekkar, H. K. Djemaa, and T. A. J. J. I. E. A. Alitouche, "Evaluation measures for models assessment over imbalanced data sets," in Journal of Information Engineering and Applications, 2013.
34- W. J. J. C. Youden, "Index for rating diagnostic tests," in Cancer, pp. 32-35, 1950.
35- S. Brin, R. Motwani, J. D. Ullman, and S. J. A. S. R. Tsur, "Dynamic itemset counting and implication rules for market basket data," in Proceedings of the 1997 ACM SIGMOD international conference on Management of datavol, pp. 255-264, 1997.
36- S. J. T. J. o. O. S. S. Raschka, "MLxtend: Providing machine learning and data science utilities and extensions to Python’s scientific computing stack," in Journal of Open Source Software, 2018.
37- J. BRAy and J. CuRTIS, "An ordination of upland forest communities of southern Wisconsin.-ecological Monographs," in journal of Ecological Monographs, 1957.
38- J. L. Fleiss, J. Cohen, and B. S. J. P. B. Everitt, "Large sample standard errors of kappa and weighted kappa," in Psychological Bulletin, p. 323, 1969.
39- M. Felkin, "Comparing classification results between n-ary and binary problems," in Quality Measures in Data Mining: Springer, pp. 277-301, 2007.
40- R. Ranawana and V. Palade, "Optimized Precision-A new measure for classifier performance evaluation," in 2006 IEEE International Conference on Evolutionary Computation, pp. 2254-2261, 2006.
41- V. García, R. A. Mollineda, and J. S. Sánchez, "Index of balanced accuracy: A performance measure for skewed class distributions," in Iberian Conference on Pattern Recognition and Image Analysis, pp. 441-448, 2009.
42- P. Branco, L. Torgo, and R. P. J. A. C. S. Ribeiro, "A survey of predictive modeling on imbalanced domains," in Journal ACM Computing Surveys (CSUR), p. 31, 2016.
43- K. Pearson, "Notes on Regression and Inheritance in the Case of Two Parents," in Proceedings of the Royal Society of London, p. 240-242, 1895.
44- W. J. I. Conover, New York, "Practical Nonparametric Statistics," in John Wiley and Sons, 1999.
45- Yule, G. U, "On the methods of measuring association between two attributes." in Journal of the Royal Statistical Society, pp. 579-652, 1912.
46- Batuwita, R. and Palade, V, "A new performance measure for class imbalance learning. application to bioinformatics problems," in Machine Learning and Applications, pp.545–550, 2009.
47- D. K. Lee, "Alternatives to P value: confidence interval and effect size," Korean journal of anesthesiology, vol. 69, no. 6, p. 555, 2016.
48- M. A. Raslich, R. J. Markert, and S. A. Stutes, "Selecting and interpreting diagnostic tests," Biochemia medica: Biochemia medica, vol. 17, no. 2, pp. 151-161, 2007.
49- D. E. Hinkle, W. Wiersma, and S. G. Jurs, "Applied statistics for the behavioral sciences," 1988.
50- A. Maratea, A. Petrosino, and M. Manzo, "Adjusted F-measure and kernel scaling for imbalanced data learning," Information Sciences, vol. 257, pp. 331-341, 2014.
51- L. Mosley, "A balanced approach to the multi-class imbalance problem," 2013.
52- M. Vijaymeena and K. Kavitha, "A survey on similarity measures in text mining," Machine Learning and Applications: An International Journal, vol. 3, no. 2, pp. 19-28, 2016.
53- Y. Otsuka, "The faunal character of the Japanese Pleistocene marine Mollusca, as evidence of climate having become colder during the Pleistocene in Japan," Biogeograph. Soc. Japan, vol. 6, pp. 165-170, 1936.
Cite
If you use PyCM in your research, please cite this paper :
Haghighi, S., Jasemi, M., Hessabi, S. and Zolanvari, A. (2018). PyCM: Multiclass confusion matrix library in Python. Journal of Open Source Software, 3(25), p.729.
@article{Haghighi2018,
doi = {10.21105/joss.00729},
url = {https://doi.org/10.21105/joss.00729},
year = {2018},
month = {may},
publisher = {The Open Journal},
volume = {3},
number = {25},
pages = {729},
author = {Sepand Haghighi and Masoomeh Jasemi and Shaahin Hessabi and Alireza Zolanvari},
title = {{PyCM}: Multiclass confusion matrix library in Python},
journal = {Journal of Open Source Software}
}
Download PyCM.bib
| JOSS |  |
| Zenodo |  |
| Researchgate |  |
License
Donate to our project
If you do like our project and we hope that you do, can you please support us? Our project is not and is never going to be working for profit. We need the money just so we can continue doing what we do ;-) .
Changelog
All notable changes to this project will be documented in this file.
The format is based on Keep a Changelog and this project adheres to Semantic Versioning.
Unreleased
2.3 - 2019-06-27
Added
- Adjusted F-score (AGF)
- Overlap coefficient (OC)
- Otsuka-Ochiai coefficient (OOC)
Changed
save_statandsave_vectorparameters added tosave_objmethod- Document modified
README.mdmodified- Parameters recommendation for imbalance dataset modified
- Minor bug in
Compareclass fixed pycm_helpfunction modified- Benchmarks color modified
2.2 - 2019-05-30
Added
- Negative likelihood ratio interpretation (NLRI)
- Cramer's benchmark (SOA5)
- Matthews correlation coefficient interpretation (MCCI)
- Matthews's benchmark (SOA6)
- F1 macro
- F1 micro
- Accuracy macro
Changed
Compareclass score calculation modified- Parameters recommendation for multi-class dataset modified
- Parameters recommendation for imbalance dataset modified
README.mdmodified- Document modified
- Logo updated
2.1 - 2019-05-06
Added
- Adjusted geometric mean (AGM)
- Yule's Q (Q)
Compareclass and parameters recommendation system block diagrams
Changed
- Document links bug fixed
- Document modified
2.0 - 2019-04-15
Added
- G-Mean (GM)
- Index of balanced accuracy (IBA)
- Optimized precision (OP)
- Pearson's C (C)
Compareclass- Parameters recommendation warning
ConfusionMatrixequal method
Changed
- Document modified
stat_printfunction bug fixedtable_printfunction bug fixedBetaparameter renamed tobeta(F_calcfunction &F_betamethod)- Parameters recommendation for imbalance dataset modified
normalizeparameter added tosave_htmlmethodpycm_func.pysplitted intopycm_class_func.pyandpycm_overall_func.pyvector_filter,vector_check,class_checkandmatrix_checkfunctions moved topycm_util.pyRACC_calcandRACCU_calcfunctions exception handler modified- Docstrings modified
1.9 - 2019-02-25
Added
- Automatic/Manual (AM)
- Bray-Curtis dissimilarity (BCD)
CODE_OF_CONDUCT.mdISSUE_TEMPLATE.mdPULL_REQUEST_TEMPLATE.mdCONTRIBUTING.md- X11 color names support for
save_htmlmethod - Parameters recommendation system
- Warning message for high dimension matrix print
- Interactive notebooks section (binder)
Changed
save_matrixandnormalizeparameters added tosave_csvmethodREADME.mdmodified- Document modified
ConfusionMatrix.__init__optimized- Document and examples output files moved to different folders
- Test system modified
relabelmethod bug fixed
1.8 - 2019-01-05
Added
- Lift score (LS)
version_check.py
Changed
colorparameter added tosave_htmlmethod- Error messages modified
- Document modified
- Website changed to http://www.pycm.ir
- Interpretation functions moved to
pycm_interpret.py - Utility functions moved to
pycm_util.py - Unnecessary
elseandelifremoved ==changed tois
1.7 - 2018-12-18
Added
- Gini index (GI)
- Example-7
pycm_profile.py
Changed
class_nameparameter added tostat,save_stat,save_csvandsave_htmlmethodsoverall_paramandclass_paramparameters empty list bug fixedmatrix_params_calc,matrix_params_from_tableandvector_filterfunctions optimizedoverall_MCC_calc,CEN_misclassification_calcandconvex_combinationfunctions optimized- Document modified
1.6 - 2018-12-06
Added
- AUC value interpretation (AUCI)
- Example-6
- Anaconda cloud package
Changed
overall_paramandclass_paramparameters added tostat,save_statandsave_htmlmethodsclass_paramparameter added tosave_csvmethod_removed from overall statistics namesREADME.mdmodified- Document modified
1.5 - 2018-11-26
Added
- Relative classifier information (RCI)
- Discriminator power (DP)
- Youden's index (Y)
- Discriminant power interpretation (DPI)
- Positive likelihood ratio interpretation (PLRI)
__len__methodrelabelmethod__class_stat_init__function__overall_stat_init__functionmatrixattribute as dictnormalized_matrixattribute as dictnormalized_tableattribute as dict
Changed
README.mdmodified- Document modified
LR+renamed toPLRLR-renamed toNLRnormalized_matrixmethod renamed toprint_normalized_matrixmatrixmethod renamed toprint_matrixentropy_calcfixedcross_entropy_calcfixedconditional_entropy_calcfixedprint_tablebug for large numbers fixed- JSON key bug in
save_objfixed transposebug insave_objfixedPython 3.7added to.travis.yamlandappveyor.yml
1.4 - 2018-11-12
Added
- Area under curve (AUC)
- AUNU
- AUNP
- Class balance accuracy (CBA)
- Global performance index (RR)
- Overall MCC
- Distance index (dInd)
- Similarity index (sInd)
one_vs_alldev-requirements.txt
Changed
README.mdmodified- Document modified
save_statmodifiedrequirements.txtmodified
1.3 - 2018-10-10
Added
- Confusion entropy (CEN)
- Overall confusion entropy (Overall CEN)
- Modified confusion entropy (MCEN)
- Overall modified confusion entropy (Overall MCEN)
- Information score (IS)
Changed
README.mdmodified
1.2 - 2018-10-01
Added
- No information rate (NIR)
- P-Value
sample_weighttranspose
Changed
README.mdmodified- Key error in some parameters fixed
OSXenv added to.travis.yml
1.1 - 2018-09-08
Added
- Zero-one loss
- Support
online_helpfunction
Changed
README.mdmodifiedhtml_tablefunction modifiedtable_printfunction modifiednormalized_table_printfunction modified
1.0 - 2018-08-30
Added
- Hamming loss
Changed
README.mdmodified
0.9.5 - 2018-07-08
Added
- Obj load
- Obj save
- Example-4
Changed
README.mdmodified- Block diagram updated
0.9 - 2018-06-28
Added
- Activation threshold
- Example-3
- Jaccard index
- Overall Jaccard index
Changed
README.mdmodifiedsetup.pymodified
0.8.6 - 2018-05-31
Added
- Example section in document
- Python 2.7 CI
- JOSS paper pdf
Changed
- Cite section
- ConfusionMatrix docstring
- round function changed to numpy.around
README.mdmodified
0.8.5 - 2018-05-21
Added
- Example-1 (Comparison of three different classifiers)
- Example-2 (How to plot via matplotlib)
- JOSS paper
- ConfusionMatrix docstring
Changed
- Table size in HTML report
- Test system
README.mdmodified
0.8.1 - 2018-03-22
Added
- Goodman and Kruskal's lambda B
- Goodman and Kruskal's lambda A
- Cross entropy
- Conditional entropy
- Joint entropy
- Reference entropy
- Response entropy
- Kullback-Liebler divergence
- Direct ConfusionMatrix
- Kappa unbiased
- Kappa no prevalence
- Random accuracy unbiased
pycmVectorErrorclasspycmMatrixErrorclass- Mutual information
- Support
numpyarrays
Changed
- Notebook file updated
Removed
pycmErrorclass
0.7 - 2018-02-26
Added
- Cramer's V
- 95% confidence interval
- Chi-Squared
- Phi-Squared
- Chi-Squared DF
- Standard error
- Kappa standard error
- Kappa 95% confidence interval
- Cicchetti benchmark
Changed
- Overall statistics color in HTML report
- Parameters description link in HTML report
0.6 - 2018-02-21
Added
- CSV report
- Changelog
- Output files
digitparameter toConfusionMatrixobject
Changed
- Confusion matrix color in HTML report
- Parameters description link in HTML report
- Capitalize descriptions
0.5 - 2018-02-17
Added
- Scott's pi
- Gwet's AC1
- Bennett S score
- HTML report
0.4 - 2018-02-05
Added
- TPR micro/macro
- PPV micro/macro
- Overall RACC
- Error rate (ERR)
- FBeta score
- F0.5
- F2
- Fleiss benchmark
- Altman benchmark
- Output file(.pycm)
Changed
- Class with zero item
- Normalized matrix
Removed
- Kappa and SOA for each class
0.3 - 2018-01-27
Added
- Kappa
- Random accuracy
- Landis and Koch benchmark
overall_stat
0.2 - 2018-01-24
Added
- Population
- Condition positive
- Condition negative
- Test outcome positive
- Test outcome negative
- Prevalence
- G-measure
- Matrix method
- Normalized matrix method
- Params method
Changed
statistic_resulttoclass_statparamstostat
0.1 - 2018-01-22
Added
- ACC
- BM
- DOR
- F1-Score
- FDR
- FNR
- FOR
- FPR
- LR+
- LR-
- MCC
- MK
- NPV
- PPV
- TNR
- TPR
- documents and
README.md
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pycm-2.3.tar.gz.
File metadata
- Download URL: pycm-2.3.tar.gz
- Upload date:
- Size: 721.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.12.1 pkginfo/1.4.2 requests/2.20.1 setuptools/18.5 requests-toolbelt/0.8.0 tqdm/4.28.1 CPython/3.4.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
25e49df3325b7d38ac61199eb8017a20074434b197f7242f9efb3f856e6ff5ae
|
|
| MD5 |
dd878866d31f2de7384961afa0e9bac0
|
|
| BLAKE2b-256 |
594de97ff8f9b60e00c044ca20964d95574aec9a7bd47fbf80910ea09194bbfe
|
File details
Details for the file pycm-2.3-py2.py3-none-any.whl.
File metadata
- Download URL: pycm-2.3-py2.py3-none-any.whl
- Upload date:
- Size: 47.4 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.12.1 pkginfo/1.4.2 requests/2.20.1 setuptools/18.5 requests-toolbelt/0.8.0 tqdm/4.28.1 CPython/3.4.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
6e529e246b57ca8b31a8a80e076be5720df1fd981d518d3002441257db36a92b
|
|
| MD5 |
c818e54f30ee1034acd7e6e7b190f44e
|
|
| BLAKE2b-256 |
e84463901f094f7d2ab59b598a5a3aae8bf476a1030368aefa6ada736acf9002
|